Abstract
Human prostate cancer (PCa) is known to harbor recurrent genomic aberrations consisting of chromosomal losses, gains, rearrangements and mutations that involve oncogenes and tumor suppressors. Genetically engineered mouse (GEM) models have been constructed to assess the causal role of these putative oncogenic events and provide molecular insight into disease pathogenesis. While GEM models generally initiate neoplasia by manipulating a single gene, expression profiles of GEM tumors typically comprise hundreds of transcript alterations. It is unclear whether these transcriptional changes represent the pleiotropic effects of single oncogenes, and/or cooperating genomic or epigenomic events. Therefore, it was determined if structural chromosomal alterations occur in GEM models of PCa and whether the changes are concordant with human carcinomas. Whole genome array-based comparative genomic hybridization (CGH) was used to identify somatic chromosomal copy number aberrations (SCNAs) in the widely used TRAMP, Hi-Myc, Pten-null and LADY GEM models. Interestingly, very few SCNAs were identified and the genomic architecture of Hi-Myc, Pten-null and LADY tumors were essentially identical to the germline. TRAMP neuroendocrine carcinomas contained SCNAs, which comprised three recurrent aberrations including a single copy loss of chromosome 19 (encoding Pten). In contrast, cell lines derived from the TRAMP, Hi-Myc, and Pten-null tumors were notable for numerous SCNAs that included copy gains of chromosome 15 (encoding Myc) and losses of chromosome 11 (encoding p53).
Keywords: prostate cancer, genome, mouse model, CGH, genetically engineered mouse
INTRODUCTION
To aid in evaluating genetic and environmental factors that influence the development and progression of prostate cancer, murine models have been developed that employ gene targeting technology to alter key signaling programs that regulate aspects of cellular proliferation and survival. The mouse does not naturally develop prostate cancer, and although the rodent prostate differs from the human prostate anatomically, several genetically engineered mouse (GEM) models exhibit attributes also observed in human prostate cancer. Notably, histological features of prostatic intraepithelial neoplasia (PIN), locally invasive adenocarcinoma, and neuroendocrine carcinoma are recapitulated in GEM prostate cancer models (reviewed in (1, 2)). Several models also regress following systemic androgen suppression or pharmacological inhibition of androgen receptor (AR) signaling, a hallmark of clinical responses observed in human prostate cancer.
The Transgenic Adenocarcinoma of the Mouse Prostate (TRAMP) was the first prostate cancer GEM model developed. This model expresses SV40 T-antigen (Tag) under a minimal probasin promoter which specifically targets transgene expression to the prostatic epithelium (3, 4). The full SV40 Tag abrogates the function of the retinoblastoma (RB1), TRP53, and PP2A tumor suppressor proteins (5), each of which are altered in a subset of primary human prostate cancers (6). A second T antigen GEM model, designated LADY, expresses the large T-antigen, lacks small t-antigen, and consequently inactivates only RB1 and TRP53 function. LADY transgenic mice develop murine PIN (mPIN) that progresses to adenocarcinoma (7). The tumor suppressor PTEN is lost in ~40% of human prostate tumors (6) and GEM models with prostate-specific deletion of Pten develop mPIN and ultimately invasive adenocarcinoma and metastasis at a low frequency (8). Increased copy number of the Myc locus and overexpression of MYC protein are also common in human prostate cancer (6). Mice strains engineered to overexpress MYC in prostate epithelium develop mPIN that progresses to locally invasive adenocarcinoma by 3–6 months.
In addition to the histological similarities between the neoplasms that develop spontaneously in humans and as a consequence of genetic manipulation in the mouse, cross-species comparisons of gene expression have identified syntenic downstream molecular alterations (8, 9). While concordant changes in the specific pathway perturbed in the human cancer and corresponding GEM model are anticipated, profiling studies indicate that additional alterations accompany the initiating event, and the patterns and networks of gene expression exhibit parallels with human cancers. This association has been seen in several GEM models for breast, lung, colon and prostate cancer, among others (9–12). These observations suggest that cooperating genomic and/or epigenetic events might be shared between species, which could explain in part the recurrent deregulation of a subset of key obligate genes promoting the transition of premalignant cells to invasive neoplasms.
In human carcinomas, phenotypic changes associated with gene expression can often be attributed to underlying alterations in the genomic architecture of the cancer cell—regions of DNA copy gain, DNA loss, aneuploidy, and nucleotide insertions, deletions, and base changes. Studies of primary and metastatic human prostate cancers report numerous recurrent genomic alterations: at the time of diagnosis, the genome of a typical primary prostate cancer harbors between 10–100 non-synonymous nucleotide mutations and multiple chromosomal rearrangements and somatic copy number aberrations (SCNAs) (6, 13–16). In addition to TRP53, RB, PTEN and MYC described above, examples of recurrent genomic aberrations include mutations in SPOP (~13%), MED12 (5%), rearrangements of the TMRPSS2-ERG locus (~50%), loss of chromosome 8p (~30–50%), and gain of chromosome 8q (~20–40%).
The clonal and recurrent nature of genomic aberrations in human prostate cancers strongly suggests that the genes and/or regulatory elements contained in these loci contribute to neoplastic growth. Prostate tumors rarely have a single anomaly, but rather commonly harbor multiple recurrent genomic alterations, a finding that strongly suggests a requirement for cooperating events to effectively drive malignant phenotypes. To date there is little information concerning whether recurrent genomic aberrations in GEM models of prostate cancer associate with neoplastic progression and underlie the extensive gene expression alterations observed in these models. As chromosomal structural alterations dominate the mutational landscape of human prostate cancers, we undertook this study to determine if recurrent SCNAs occur in the tumors (and derived cell lines) from GEM models of prostate cancer, to determine if these alterations associate with the specific driver events, and assess whether the genomic changes are concordant with those commonly found in human prostate cancers.
MATERIALS AND METHODS
Genetically Engineered Mouse Models
The TRAMP C57BL/6 FVB F1 mice used in these studies were generated as follows: C57BL/6 (B6) TRAMP mice were obtained from Dr. Norman Greenberg (Fred Hutchinson Cancer Center, Seattle, WA) and were subsequently bred by continued backcrossing to B6 mice (Jackson labs). FVB/NTac mice were obtained from Taconic (Germantown, NY). B6 TRAMP females were mated with FVB males to generate B6FVBF1 TRAMP animals. Hi-Myc mice were obtained from the Mouse Repository of the National Cancer Institute Mouse Models of Human Cancer Consortium. Hemizygous Hi-Myc mice on FVB background were cross-bred with non transgenic FVB breeders from Taconic (Germantown, NY). B6FVB F1 TRAMP mice between 24 to 29 weeks old and Hi-Myc mice between 56–72 weeks-old were sacrificed by cervical dislocation. Spleens were removed and snap-frozen in liquid nitrogen. Prostate glands were dissected and cut into 2 pieces, one was processed for histology and the other was snap-frozen in liquid nitrogen and stored at −80°C until DNA/RNA extraction. All animals were maintained pathogen free in the Fred Hutchinson Cancer Research Center animal facility that is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. The Pb-Cre+;PtenL/L GEM was generated and propagated in a C57BL/6. DBA.129/Balb/c background as previously described (8). This strain was maintained at the University of California Los Angeles by Dr. Hong Wu. Prostate glands from Pb-Cre+;PtenL/L at age 24 weeks were resected, snap frozen, and processed as previously described (8).
Cell lines derived from GEM models of prostate cancer
TRAMP-C2 cells were a generous gift from Dr. Norman Greenberg (Fred Hutchinson Cancer Research Center, Seattle, WA) and were originally derived from a primary prostate tumor of a 32 week-old PB-Tag C57BL/6 (TRAMP) mouse (17). Myc-CaP cells were established from a primary prostate carcinoma dissected from a 16-month-old Hi-Myc transgenic mouse in the FVB inbred strain (18). Myc-CaP and TRAMP-C2 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C. Pten-P8 cells were established from a primary prostate tumor dissected from a 10-month-old Pb-Cre+;PtenL/L mouse (19). Pten-P8 were maintained in DMEM supplemented with 10% fetal bovine serum (FBS; Omega Scientific), 25 Ag/mL bovine pituitary extract, 5 Ag/mL bovine insulin, and 6 ng/mL recombinant human epidermal growth factor (Sigma-Aldrich). Cell lines were authenticated within 3 passages of performing CGH studies by comparing gene expression profiles with previously published expression profiles and confirming concordant relationships by unsupervised clustering.
RNA and DNA extraction
Approximately 5 mg of frozen prostate tumor or spleen were ground in liquid nitrogen containing 350 µl of RLT plus buffer, and the resulting powder was placed in Eppendorf tubes and store at −80C until DNA/RNA extractions. DNA and RNA extraction was done using the Qiagen All Prep DNA/RNA kit (QIAGEN, USA) according to the manufacturer’s instructions. The reference DNA (wild-type male C57BL/6 mouse) was purchased from The Jackson Laboratory (Bar Harbor, ME).
Copy number analysis by array Comparative Genomic Hybridization
Paraffin-embedded (TRAMP samples) or O.C.T embedded (Hi-Myc and Pb-Cre+;PtenL/L) prostate tumor sections were stained with hematoxylin and eosin according to standard protocols. All tissues were cut into 4 or 5µM sections and stained with hematoxylin and eosin (H&E) for histologic examination to confirm the presence of neoplastic cells. DNA from 3 prostate tumors and germline spleen DNA from 3 different mice, for each GEM model, and a common reference male C57BL/6 DNA were used for CGH array analysis using the Agilent Sure Print G3 mouse CGH 4 × 180K microarray platform (Agilent Technologies, Santa Clara, CA, USA). The sample preparation and hybridization protocols recommended by Agilent were followed. Briefly, we used 1.5 µg of tumor or germline spleen DNA and 1.5 µg of reference DNA for each analysis. DNA was digested with Rsa I and Alu I and labeled by random priming using either Cy3-dUTP or Cy5-dUTP, respectively. Unincorporated nucleotides were removed using Am icon Ultra-0.5 ml, Ultracel-30 membrane (Millipore), and tumor (or germline) and reference samples were combined. Probes were denatured and pre-annealed with 50 µg of mouse Cot-1 DNA (Invitrogen, USA), and 71 µl of hybridization master mix was added before loading into array slides. Hybridization was performed at 65 °C for 24 hrs at 20 rpm. After hybridization, slides were washed and scanned immediately with a DNA Microarray Scanner (Agilent Technologies). Data were extracted from scanned images using Feature Extraction software (Agilent). The text files were then imported for analysis into Genomic Workbench, standard edition 7.0.4.0 (Agilent). We used the Aberration Detection Method 2 (ADM-2) algorithm to identify DNA copy number aberrations. The statistical score represents the deviation of the average of the log ratios from the expected value of zero, in units of standard deviation. The following parameters were used in this analysis: Threshold of ADM-2: 7.0; Fuzzy Zero: ON; GC correction: ON, Diploid Peak Centralization: ON. Aberration Filters: ON (Minimum Number of Probes for Amplification > = 6 AND Minimum Size (Kb) of Region for Amplification > = 0.0 AND Minimum Avg. Absolute Log Ratio for Amplification > = 0.25) OR (Minimum Number of Probes for Deletion > = 6 AND Minimum Size (Kb) of Region for Deletion > = 0.0 AND Minimum Avg. Absolute Log Ratio for Deletion > = 0.25). Genomic positions were based on the UCSC July 2007 mouse reference sequence (NCBI37/mm9).
RESULTS
Genomic aberrations in the TRAMP GEM model
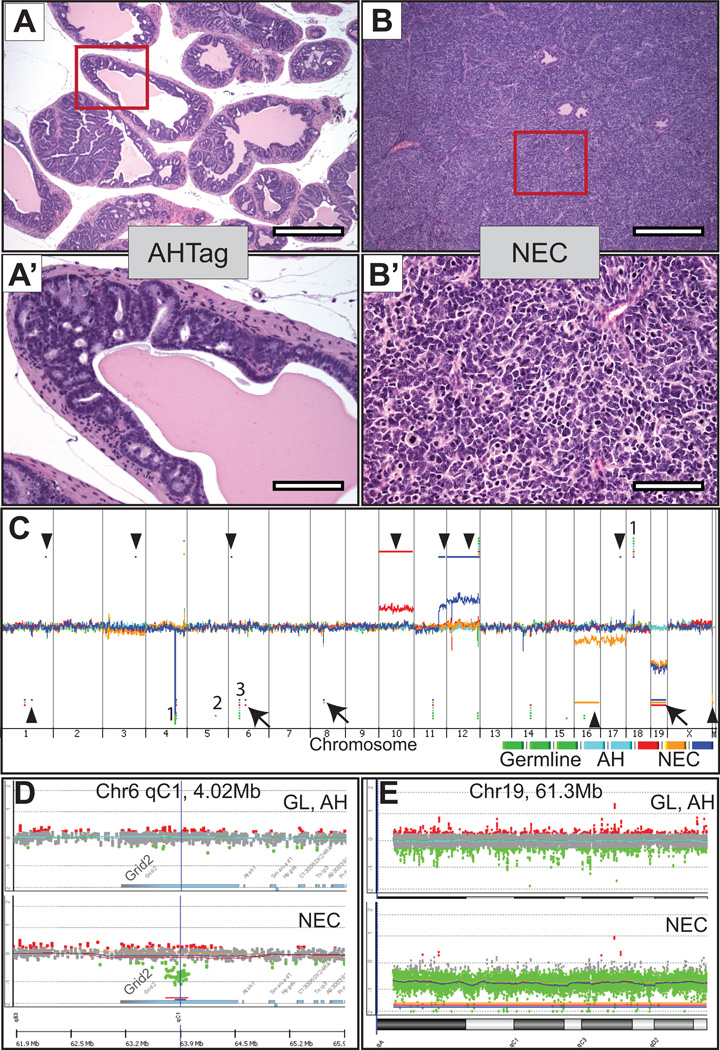
In the TRAMP model, the onset of prostate tumor development varies depending on the strain background (3, 20). In this study, we evaluated prostatic neoplasms arising in a mixed C57BL/6 × FVB/N background as the FVB genotype exhibits reduced latency, enhanced primary tumor growth and high rates of metastasis when compared to tumor development in a pure C57BL/6 background (3, 20). We resected the prostate glands from 24–29 week-old TRAMP [C57BL/6×FVB] F1 male mice and analyzed three prostate neuroendocrine carcinomas (NEC, n = 3), two prostates (from littermate mice) with atypical hyperplasia of Tag (AHTag), and used spleen DNA from three independent mice as the reference to control for strain-specific genomic variation. DNA from AHTag and NE carcinomas were extracted from a defined tumor mass where at least 70% of the region contained hyperplastic/NE cells, as determined by hematoxylin and eosin stains (Figure 1A and Figure 1B, respectively).
Figure 1. Histology and genomic copy number alterations in TRAMP tumors.
A–B’ Representative microscopic images from hematoxylin and eosin-stained TRAMP sections from the prostate glands from which DNA was extracted for CGH analysis. A-A’, TRAMP atypical hyperplasia of Tag (AHTag) at low (4×) and high-power (20×) respectively. B-B’, TRAMP neuroendocrine carcinoma (NEC) at low (4×) and high-power (20×) respectively. C, Copy number alterations (CNAs) in TRAMP mice by array CGH. Genomic DNA from C57BL/6. FVB TRAMP prostate NE carcinomas, AHTag lesions and spleen germline DNA control were hybridized against sex-matched normal mouse C57BL/6 reference DNA using Agilent CGH slides containing 180K probes. Whole genome view of overlaid moving averages (2 Mb window) for the log2 ratios of fluorescence between a sample/reference DNA probe (Y axis) plotted at its genomic position (X axis), red, blue and yellow (NE tumors; n = 3), cyan blue shades (AHTag n = 2) and green shades (germline n = 3). Aberrations called in any samples (germline and tumor) by the ADM-2 algorithm are identified by a horizontal bar (red, blue and yellow: NE tumors), Cyan (AHTag) and Green (germline DNA). Aberrations present in at least two (out of 3) NE tumors are indicated by arrows; those found in individual mice are indicated by arrowheads. (1) Representative CNVs in common between germline, AHTag and NE tumors; (2) Aberrations called by the ADM-2 algorithm in a single sample that by manual inspection exhibited similar fluorescence intensities in all samples including germline and tumor (see Supplemental Figure S2, for representative examples); (3) Example of a known strain-specific alteration in the FVB strain (a copy number loss in the t-cell receptor V-beta gene segment (chr6:41,018,389–41,112,757). D, Zoom-in view of chromosome 6 showing overlaid data points for log2 ratios of the Grid2 gene region. Green: values below log2 = −0.25; Red: values above log2 = 0.25. Upper panel spleen and AHTag samples. Lower panel NE tumors showing a SCNAs in the Grid 2 gene. Aberrations called by the ADM-2 algorithm are identified by a horizontal line (bottom). E, Zoom-in on whole chromosome 19 showing overlaid data points for log2 ratios. Green: values below log2 = −0.25; Red: values above log2 = 0.25. Upper panel spleen and AHTag samples. Lower panel NE tumors showing a copy number loss of whole chr19. The aberration called by the ADM-2 algorithm of the entire chromosome 19 is identified by a horizontal line (bottom).
A comparison of array CGH signals across the genomes of TRAMP mice and the C57BL/6 reference DNA clearly identified strain background-associated copy number variance (CNVs), indicating FVB specific CNVs such as a copy number loss in the t-cell receptor V-beta gene segment (chr6:41,018,389–41,112,757), typical of the FVB strain consistent with previous reports (21, 22) (Figure 1C).
In prostate neoplasms, the genomes derived from AHTag lesions did not exhibit any SCNAs (Figure 1C, cyan-blue). However, between 3 and 10 SCNAs were identified in neuroendocrine (NE) carcinomas (Fig. 1C and supporting Table S1), though most were not present in more than one tumor. Gains of whole chromosomes 10, 12 and distal chromosome 11, focal gains in chromosome 1, 3 and 6, heterozygous loss of whole chromosome 16 and focal losses in chromosomes 1, 14 and Y were notable in individual tumors (Figure 1C). Only three regions were shared in at least two of the 3 NE tumors tested. The first was a localized heterozygous deletion (0.13 Mb segment,) on chromosome 6 (chr6:63,863,932–63,996,654), a map location for the Grid2 gene (Figure 1D). The second was a 0.68Mbdeletion on chromosome 8 (chr8: 50,762,466–51,279,003). The third region was a heterozygous copy number loss of the entire chromosome 19 (the location of murine Pten) observed in each of the NE carcinomas analyzed (Figure 1C,E). Of interest, 24% (9out of 37 cases) of metastatic human prostate cancers evaluated by array CGH exhibited heterozygous loss of the syntenic human genomic region on chromosome 4 encompassing GRID2, (chr4:93,225,550–94,693,649)(Supplementary Figure S1) (6). However, GRID2 transcript levels were not significantly reduced in human or mouse tumors with heterozygous GRID2 loss (Supplementary Figure S1 and S2).
To determine whether the loss of one chromosome 19 copy results in an overall down-regulation of the genes located on this chromosome, we evaluated the corresponding transcript levels by gene expression arrays from an independent sample group. Overall, transcript levels for genes located on chromosome 19 were lower in TRAMP tumors compared to the normal tissues (p<0.001) (Figure S2). However, transcripts encoding the tumor suppressor gene Pten, located in chromosome 19 were not significantly decreased in TRAMP NE tumors. This is consistent with a previous study of TRAMP tumors reporting the loss of one Pten allele in 50% of the mice, but no decrease in PTEN protein was observed, nor were inactivating mutations in the other Pten allele detected (23). In contrast, transcripts encoding the MEN1 tumor suppressor were diminished in tumors relative to benign prostate (p<0.05) (Supplementary Figure S2). Of interest, mutations in MEN1 confer genetic predisposition to neuroendocrine tumors in humans (24) and a subset of heterozygous Men1+/− mice are reported to develop prostate pathologies in advanced age (25).
We also evaluated the genomic integrity of tumors obtained from the LADYGEM model (line LPB-Tag 12T-7f) which exhibits several features also found in the TRAMP GEM model such as the development of neuroendocrine tumors. This strain typically develops hyperplasia and carcinomas by 21 weeks of age. No SCNAs were identified in the tumors resected from 21-week old LPB-Tag 12T-7f mice (see Supplementary Figure S3 and Table S2).
Genomic aberrations in the Pten-null GEM model
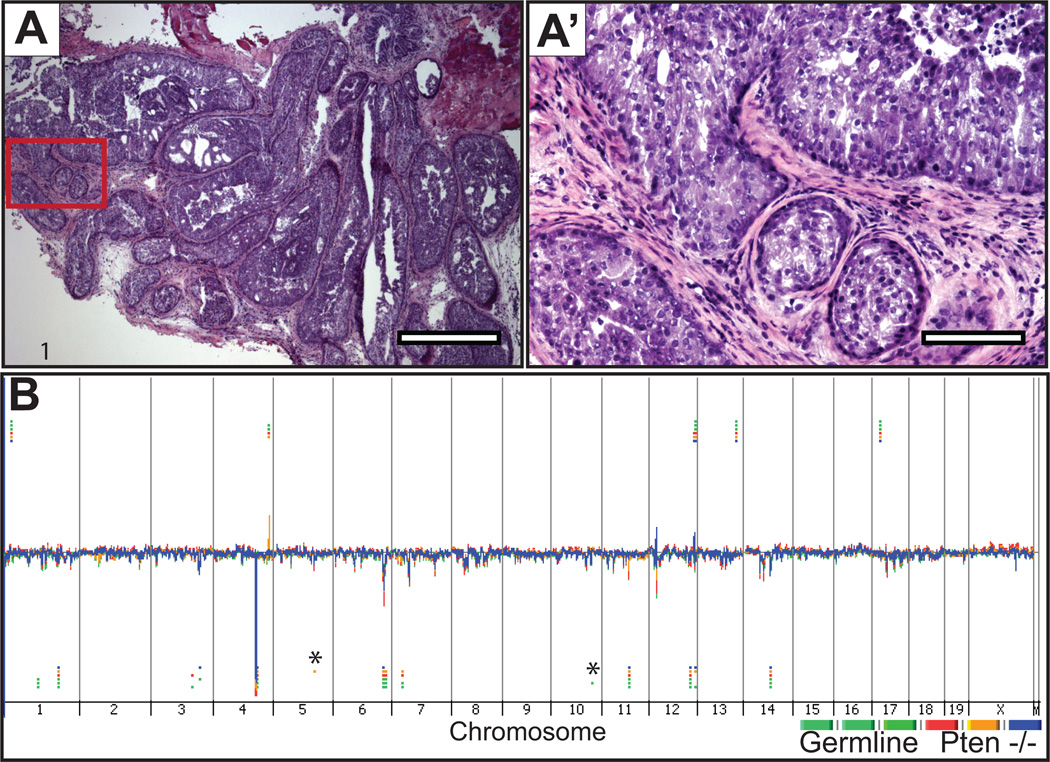
In the Pb-Cre;PtenL/L model, high grade intraepithelial neoplasia or invasive carcinomas are evident by 24 weeks of age (8). We resected prostate glands and the spleens from 24 week Pb-Cre;PtenL/L mice and extracted DNA from whole Pten-null prostates after confirming that≥ 70% of each sample comprised neoplastic cells (See Figure 2A-A’). At the genomic level, Pten-null prostate tumors did not exhibit any unique somatic copy number changes identifiable by CGH (Figure 2B). All CNAs were present in both germline and tumor tissue DNA, except one copy number loss (chr5:105142362–105237345) called by the ADM-2 algorithm that appeared to be unique to one Pten tumor (asterisk in Figure 2B). However, manual inspection of the probes’ intensity levels for that region revealed similar levels across all tumors and germline samples (see Supplemental Figure S4 and Table S3). In order to call aberrations, we required a minimum number of 6 probes to call an amplification or deletion. The CNA identified to be unique to one Pten-null tumor comprised only 6 probes in the region. The other germline and tumor samples exhibited overall similar intensity levels in the region, but did not contain the minimum number of probes to call an aberration. Likewise two additional copy number losses that appeared unique to the germline control samples demonstrated similar intensity levels across tumors and germline DNA. Thus, these unique calls are likely to represent germ-line copy number variation instead of somatic copy number changes.
Figure 2. Histology and genomic view of prostates from Pten null (Pb-Cre+;PtenL/L) mice.
A-A’, representative microscopic images from hematoxylin and eosin-stained sections of prostate glands from which DNA was extracted for CGH hybridization shown at low (4×) and high-power (20×) magnification, respectively. B, Genomic DNA from Pb-Cre+;PtenL/L prostate tumors and germline spleen were hybridized against sex-matched normal mouse C57BL/6 reference DNA using Agilent CGH slides containing 180K probes. Whole genome view of overlaid moving averages (2 Mb window) for the log2 ratios of fluorescence between a sample/reference DNA probe (Y axis) plotted at its genomic position (X axis), red, blue and yellow (tumors; n = 3) and green shades (germline n = 3). Aberrations called by the ADM-2 algorithm are identified by horizontal bars. (*) Aberrations called by the ADM-2 algorithm in a single sample, that by manual inspection exhibited similar fluorescence intensities in all samples including germline and tumor (see Supplemental Figure S4, for representative examples). (1) CNAs in common between germline, and Pten lesions that are not visually obvious in the genome view window.
Genomic aberrations in the Hi-Myc GEM model
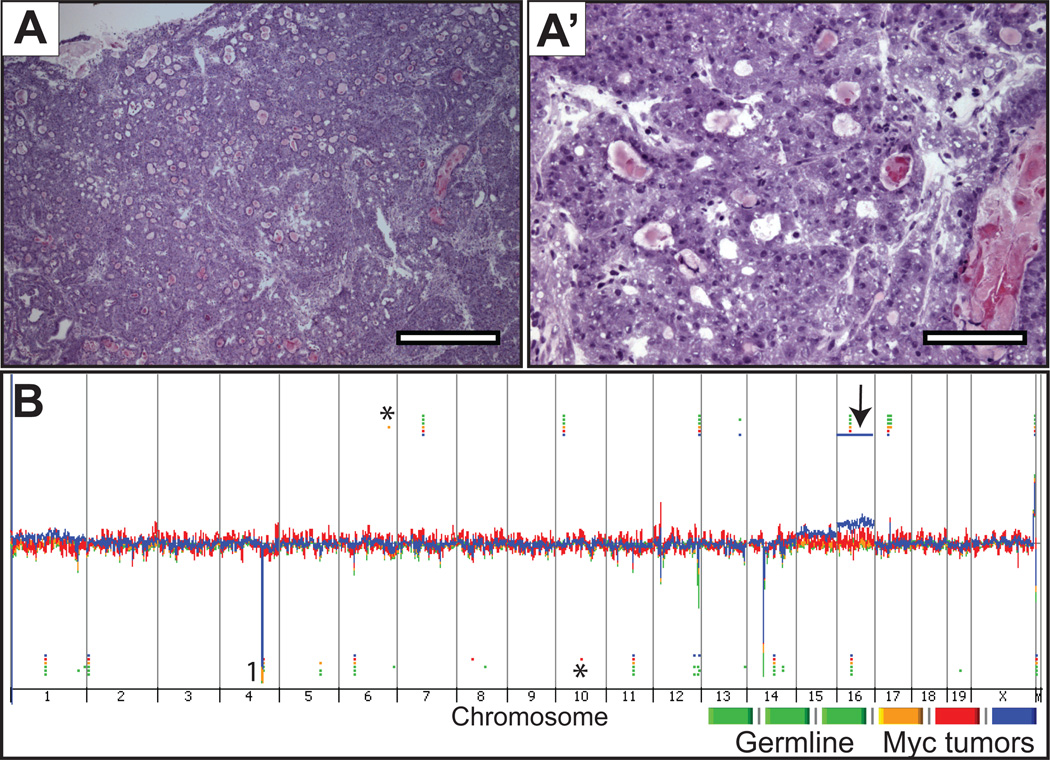
The Hi-Myc GEM model is notable for the consistent development of mPIN at 2–12 weeks and carcinoma by 24 weeks. We resected the prostates and spleens from each of 3 ARR2/probasin-Myc transgenic mice aged 56–84 weeks and confirmed that the prostate tumor regions contained ≥70% neoplastic cells (Figure 3A). The Hi-Myc mice developed a distinctive tumor mass that could be readily resected from other areas of normal prostate. Array CGH analysis of three prostate carcinomas demonstrated no recurrent regions of copy number losses or gains. One unique SCNA was identified in each tumor (see Figure 3B): a focal gain within chromosome 6 (chr6: 129795763–129841165) was present in Tumor 1; Tumor 2 exhibited a localized loss within chromosome 10 (chr10: 67536983–67741827, Arid5b) and a complete gain of chromosome 16 (Figure 3B, arrow) was present in Tumor 3. Manual inspection of the probe intensity for each of these 3 CNAs showed similar intensities across all germline and tumor samples for the focal gain (chr6) and loss (chr10) (see Supplemental Figure S5B and C), indicating the presence of a copy number variant instead of a somatic genomic aberration. The gain of whole chromosome 16 (Supplementary Figure S5A) appears to be a true SCNA. See Table S4 for complete list of CNAs called by the ADM-2 algorithm.
Figure 3. Histology and genome view of c-Myc prostate tumors.
A-A’, representative microscopic images from hematoxylin and eosin-stained sections from frozen prostate tissue where the DNA was extracted for CGH hybridization, at low (4×) and high-power (20×) magnification, respectively. B, Genomic DNA from c-Myc prostate tumors and spleen were hybridized against sex-matched normal mouse C57BL/6 reference DNA using Agilent CGH slides containing 180K probes. Whole genome view of overlaid moving averages (2 Mb window) for the log2 ratios of fluorescence between a sample (germline and c-Myc tumor DNA) and reference (sex-matched normal mouse C57BL/6) DNA probe (Y axis) plotted at its genomic position (X axis). Red, blue and yellow (tumors; n = 3) and green shades (germline n = 3). Aberrations called by the ADM-2 algorithm are identified by horizontal bars. (*) Aberrations called by the ADM-2 algorithm in single samples, that by manual inspection show similar fluorescence intensities in all samples including germline and tumor (see supplemental Figure S5, for representative examples); (1) CNAs in common between germline and Hi-Myc tumors that are not visually obvious in the genome view window.
Genomic aberrations in cell lines established from TRAMP, Pten-null and Hi-Myc GEM prostate cancer models
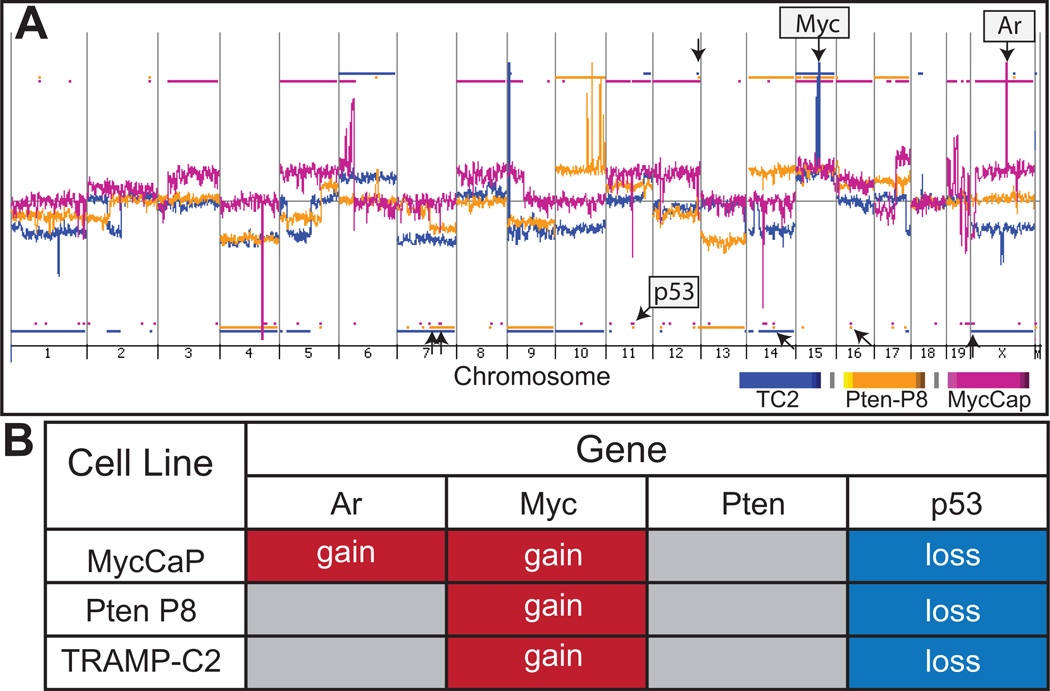
Immortal cell lines from the TRAMP, Pb-Cre;PtenL/L, and ARR2PB-Myc GEM models designated TRAMP-C2, Pten-P8 and Myc-CaP, respectively, have been established and previously characterized (Table 1). We sought to determine if genomic aberrations identified in the in situ tumors of the GEM models were also found in the derived cell lines, and if new aberrations occurred during the selection and adaptation of tumor cells to in vitro growth conditions. CGH assays identified multiple genomic aberrations in the three cell lines (Figure 4). The Myc-CaP line exhibited the greatest number of SCNAs (total of 82 CNAs, with 29 gains and 53 losses) including complete gains of chromosomes 3, 5, 8, 11, 12, 15, 16 and X; Partial gain of chromosomes 6,9,17 and 19 and partial loss of chromosome 19, and several focal-copy number gains and losses, including high-level amplification of the androgen receptor (Ar) locus (see Figure 4, fuchsia) as reported previously (18). The TRAMP-C2 line (Figure 4, blue) and Pten-P8 line (Figure 4, yellow) had fewer CNAs (31 CNAs: 18 gains and 13 losses; and 31 CNAs: 15 gains and 16 losses, respectively), but these far exceeded the alterations found in the primary tumors from the corresponding GEM strains. The genomes of TRAMP-C2 cells (blue) were notable for gains of whole chromosome 3 and 15, and losses of chromosomes 1, 7, 9, 10, 14 and X, as well as partial losses of chromosome 2 and 5 (Figure 4). We did not observe the loss of chromosome 19 or the localized loss in chromosome 6 in the TRAMP-C2 cell line identified in the tissues samples from TRAMP NE carcinomas. The Pten-P8 cell line (Figure 4, yellow) exhibited gains of chromosomes 10, 14, 15 and 17; whole-chromosome losses of chromosomes 4, 9 and 13, as well as partial loss of chromosome 7 (Figure 4A (arrows) and Table S5 for complete list of CNAs). Overall, the cell line genomes were notable for alterations in regions harboring several well-studied oncogenes and tumor suppressor genes (Figure 4B). These included amplification of the Myc locus and deletion of Trp53 in each of the three cell lines.
Table 1.
Genetically engineered mouse models of prostate cancer (and derived cell lines) assessed in this study
| GEM Prostate Cancer Models | ||||
| GEM Model | Genotype strain background | Reported age at tumor onset | Age (sample collection)* | Ref. |
| TRAMP | rPB-SV40Tag [C57BL/6×.FVB] F1 | 16 weeks hyperplasia/carcinoma | 24–29 weeks | (3, 20) |
| Pten | ARR2Pb-Cre;Ptenflox/flox C57BL/6.DBA.129/Balb/c | 9–29 wks invasive carcinoma | 24 weeks | (8) |
| Hi-Myc | ARR2PB-Myc-PAI FVB | ≥26 wks invasive carcinoma | ≥ 56 weeks | (9) |
| GEM Prostate Cancer Model-Derived Cell lines | ||||
| Cell line | Mouse Source Genotype/strain background | Mouse Source (Age) | Notes | Ref. |
| TRAMP-C2 | rPB-SV40Tag C57BL/6 | 32 weeks | Lost SV40 | (17) |
| Pten-P8 | ARR2Pb-Cre;Ptenflox/flox C57BL/6.DBA.129/Balb/c | 43 weeks | heterozygous for Pten | (19) |
| Hi-Myc | ARR2 PB-Myc FVB | 70 weeks | AR amplification | (18) |
Age at which mice were sacrificed for tissue collection.
Figure 4. Copy number alterations (CNAs) in TRAMP-C2, Myc-CaP and Pten P8 cell lines.
A)Whole genome view of overlaid moving averages (2 Mb window) for the log2 ratios of fluorescence between a cell line versus the reference (sex-matched normal C57BL/6) DNA probe (Y axis) plotted at its genomic position (X axis). Blue (TRAMP-C2), fuchsia (Myc-CaP) and yellow (Pten P8). Aberrations called by the ADM-2 algorithm are identified by a shaded area, and the presence of CNAs is indicated with blue (TRAMP-C2), fuchsia (Myc-CaP) and yellow (Pten P8) horizontal bars. Aberrations present in the three cell lines are indicated by arrows. B)In all cell lines there is a gain in chromosome 15 and loss in chromosome 11, where Myc and Trp53 are encoded, respectively. The Myc-CaP cell line has a copy number gain in the X-chromosome that includes the Ar locus.
DISCUSSION
In this study, we examined the genomic integrity of tumors derived from four commonly studied GEM prostate cancer models: TRAMP, Hi-Myc, Pten-null (Pb-Cre+;PtenL/L), and LADY. Very few somatic copy-number alterations were apparent across these four GEM models: tumors from Hi-Myc, Pb-Cre+;PtenL/L and LADY GEMs demonstrated identical genomic architecture when compared to corresponding germline DNA, with the exception of one Hi-Myc tumor with a somatic gain of chromosome 16. Using a lower resolution array a previous study reported essentially no genomic aberrations in mPIN lesions and carcinomas from the Hi-Myc model, consistent with our observations using a higher resolution platform (9). The TRAMP neuroendocrine carcinoma was the only tumor type that contained SCNAs but these were few and only three recurrent aberrations were identified. Kwabi-Addo et al have previously demonstrated loss of heterozygosity (LOH) near the Pten locus in almost 50% of TRAMP (C57BL/6) tumors, but found no evidence that the retained Pten allele was inactivated (23). The reported LOH near the Pten locus in TRAMP tumors may reflect loss of the entire chromosome 19 as observed in our study. Of interest, a recent report characterizing genomic events in murine models of small cell lung carcinoma initiated by loss of Trp53 and Rb1 also found high rates of Chr19 loss and Pten mutations (26), implicating Pten signaling as a common factor in the development of small cell carcinomas. In contrast to our assessments showing no SCNAs in Lady 12T-7f GEM tumors, an allograft model of prostate cancer that originated from a Lady 12T-10 transgenic line (NE-10) showed copy number alterations in metastatic and non-metastatic grafts (27). The NE-10 line was established from a prostate tumor of a 12T-10 transgenic mouse implanted subcutaneously and serially passaged in male athymic nude mice. Several of the genomic aberrations reported in the NE-10 allograft tumors were also observed in our analyses of the TRAMP-C2, Myc-CaP and Pten-P8 cell lines.
There are several reports describing genomic analyses of GEM models of other malignancies. Most studies, including those of breast, lung, liver, melanoma, neuroblastoma, pancreas and colon cancer-models describe relatively few structural and copy number aberrations (28–36). Interestingly, the greatest frequency of genomic alterations have been identified in short-term cultures of primary tumors, mice with telomere dysfunction, or compound models (29, 35, 37–45). The discrepancy between highly unstable human cancer genomes compared to the almost intact genomes observed in the GEM prostate cancer models is striking. Somatic copy-number alterations are a common, if not universal, feature of human cancers (46) and the vast majority of prostate cancers harbor multiple recurrent genomic aberrations. A key difference that appears to underlie this cross-species difference in cancer genotypes centers on chromosome biology and structure, particularly telomeres (47). When the mouse genome is engineered to experience telomere dysfunction leading to shortened telomere length, genome instability is evident and tumors acquired a more human cancer-like genome with complex rearrangement and alterations, a subset of which are syntenic to loci altered in human cancers (32, 39, 48). Bojovicet al. demonstrated that short telomeres in a breast cancer transgenic model (MMTV-Neu) dramatically increased lung metastasis which correlated with specific alterations in DNA copy number and gene expression (32). Using a murine prostate cancer model driven by Pten loss and p53 deletions, Ding et al. demonstrated that telomerase dysfunction early in disease onset creates genomic aberrations crucial for telomerase-driven prostate cancer metastasis. Importantly, recurrent structural chromosomal alterations were observed that corresponded to alterations identified in human prostate cancer (39).
In striking contrast to the in vivo situation, we found that cell lines derived from tumors with stable genomes exhibited substantial genomic structural changes when propagated in vitro. The chromosomal changes observed in these cell lines are consistent with previous reports. For instance Pten-P8 has been shown to contain near 6N chromosomes with 113 to 125 chromosomes identified by SKY and G-banding tests (19). AR amplification has been observed in the Myc-CaP cell line (18). Surprisingly, a number of the chromosomal changes observed in these murine prostate cancer cell lines such as loss of chromosomes 4,9 and gain of chromosomes 11 and 15 are found in immortalized murine cell lines derived from normal kidney or bladder tissue (49). Clearly the selective pressures are different for cells growing in laboratory culture than for cells dividing in the context of a tissue with physiologic tumor microenvironment.
Cross-species transcriptome analyses have shown that GEM cancer models and human tumors exhibit significant similarities in gene expression. For example, concordant with changes found in human prostate cancers, Pten-null murine prostate cancers exhibit alterations in the expression of more than 1,000 transcripts including higher levels of clusterin, prostate stem cell antigen, and osteopontin with concomitant down regulation of the Nk×3.1 tumor suppressor (8). Similarly, signatures of transcript profiles generated from the GEM model of Myc-driven prostate cancer classifies subsets of human prostate cancers into Myc-like and non-Myc-like tumors comprised of genes such as Pim-1 with known roles in cooperating with Myc to drive tumorigenesis (9). The lack of structural genomic alterations in these and other GEM tumors indicates that complex pathway interactions are activated and potentially maintained bya single, but robust oncogenic event. The mechanisms underlying the complex gene regulatory alterations that occur in the context of minimal genomic perturbations is unclear. Epigenetic changes could contribute to this complexity. However, it has been recently shown that in contrast to their human counterparts, tumors from GEM models of medulloblastoma, Burkitt lymphoma and breast cancer harbor very few loci with DNA hypermethylation (50).
This study clearly demonstrates that gross structural chromosomal alterations are not a prerequisite for tumor formation in GEM models of prostate cancer and that secondary cooperating events do not appear to be activated by the mechanisms observed in human prostate cancer, at least during the lifespan of a typical mouse. We cannot rule out that somatic single nucleotide mutations or small insertions or deletions may occur as secondary cooperating events in these murine tumors, but in contrast to structural alterations, recurrent aberrations of these types are not a common feature of primary human prostate cancers. Newer models engineering additional alterations into the backgrounds of Pten and p53 loss may provide insights into how these events cooperate, though the fact that these and other changes do not occur de novo suggests fundamental species differences in the genomes and the micro-and macro-environments that influence tumorigenesis. Further, the stable genomes of mouse models may influence the high response rates of tumors arising in GEM cancer models to cancer-directed therapeutics, and suggests that efforts designed to engineer mouse genomes with a greater degree of instability may result in models that better reflect the biology and treatment responses found in human prostate cancer.
Supplementary Material
Implications.
Chromosomal alterations are not a prerequisite for tumor formation in GEM prostate cancer models and cooperating events do not naturally occur by mechanisms that recapitulate changes in genomic integrity as observed in human prostate cancer.
ACKNOWLEDGEMENTS
We thank colleagues in the Nelson laboratory for suggestions and comments. We thank Angelo DeMarzo, Michael Ittmann and Sue Knoblaugh for pathology assessments and insightful comments and suggestions and Ilsa Coleman for assistance with data analysis. DBF was supported by a post-doctoral training award from the Department of Defense (PC110970). This work was supported by awards from the Prostate Cancer Foundation (to HW and PSN), UO1 CA164188 to HW and PSN; R01 CA165573 and the Pacific Northwest Prostate Cancer SPORE, P50CA097186.
Footnotes
Conflict of interest disclosure statement: The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Irshad S, Abate-Shen C. Modeling prostate cancer in mice: something old, something new, something premalignant, something metastatic. Cancer and Metastasis Reviews. 2012;32:109–122. doi: 10.1007/s10555-012-9409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R, et al. Animal Models of Human Prostate Cancer: The Consensus Report of the New York Meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res. 2013;73:2718–2736. doi: 10.1158/0008-5472.CAN-12-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gingrich JR, Barrios RJ, Foster BA, Greenberg NM. Pathologic progression of autochthonous prostate cancer in the TRAMP model. Prostate cancer and prostatic diseases. 1999;2:70–75. doi: 10.1038/sj.pcan.4500296. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahuja D, Saenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24:7729–7745. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- 6.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasper S, Sheppard PC, Yan Y, Pettigrew N, Borowsky AD, Prins GS, et al. Development, progression, and androgen-dependence of prostate tumors in probasin-large T antigen transgenic mice: a model for prostate cancer. Lab Invest. 1998;78:i–xv. [PubMed] [Google Scholar]
- 8.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 9.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 10.Pfefferle AD, Herschkowitz JI, Usary J, Harrell JC, Spike BT, Adams JR, et al. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome biology. 2013;14:R125. doi: 10.1186/gb-2013-14-11-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012;72:1878–1889. doi: 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 13.Baca Sylvan C, Prandi D, Lawrence Michael S, Mosquera Juan M, Romanel A, Drier Y, et al. Punctuated Evolution of Prostate Cancer Genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat J-P, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature Genetics. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grasso CS, Wu Y-M, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 17.Foster BA, Gingrich JR, Kwon ED, Madias C, Greenberg NM. Characterization of prostatic epithelial cell lines derived from transgenic adenocarcinoma of the mouse prostate (TRAMP) model. Cancer Res. 1997;57:3325–3330. [PubMed] [Google Scholar]
- 18.Watson PA. Context-Dependent Hormone-Refractory Progression Revealed through Characterization of a Novel Murine Prostate Cancer Cell Line. Cancer Res. 2005;65:11565–11571. doi: 10.1158/0008-5472.CAN-05-3441. [DOI] [PubMed] [Google Scholar]
- 19.Jiao J, Wang S, Qiao R, Vivanco I, Watson PA, Sawyers CL, et al. Murine cell lines derived from Pten null prostate cancer show the critical role of PTEN in hormone refractory prostate cancer development. Cancer Res. 2007;67:6083–6091. doi: 10.1158/0008-5472.CAN-06-4202. [DOI] [PubMed] [Google Scholar]
- 20.Chiaverotti T, Couto SS, Donjacour A, Mao J-H, Nagase H, Cardiff RD, et al. Dissociation of Epithelial and Neuroendocrine Carcinoma Lineages in the Transgenic Adenocarcinoma of Mouse Prostate Model of Prostate Cancer. The American journal of pathology. 2008;172:236–246. doi: 10.2353/ajpath.2008.070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cutler G, Marshall LA, Chin N, Baribault H, Kassner PD. Significant gene content variation characterizes the genomes of inbred mouse strains. Genome research. 2007;17:1743–1754. doi: 10.1101/gr.6754607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, Eis PS, et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwabi-Addo B, Giri D, Schmidt K, Podsypanina K, Parsons R, Greenberg N, et al. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proceedings of the National Academy of Sciences. 2001;98:11563–11568. doi: 10.1073/pnas.201167798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barakat MT, Meeran K, Bloom SR. Neuroendocrine tumours. Endocrine-related cancer. 2004;11:1–18. doi: 10.1677/erc.0.0110001. [DOI] [PubMed] [Google Scholar]
- 25.Seigne C, Fontaniere S, Carreira C, Lu J, Tong WM, Fontaniere B, et al. Characterisation of prostate cancer lesions in heterozygous Men1 mutant mice. BMC Cancer. 2010;10:395. doi: 10.1186/1471-2407-10-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, Stewart C, Carter SL, Cibulskis K, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 2014;156:1298–1311. doi: 10.1016/j.cell.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi Y, Nandana S, Case T, Nelson C, Radmilovic T, Matusik RJ, et al. Candidate metastasis suppressor genes uncovered by array comparative genomic hybridization in a mouse allograft model of prostate cancer. Molecular Cytogenetics. 2009;2:18. doi: 10.1186/1755-8166-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon RR, Merrill ML, Hunter KW, Sørensen P, Threadgill DW, Pomp D. Dietary fat-dependent transcriptional architecture and copy number alterations associated with modifiers of mammary cancer metastasis. Clinical & Experimental Metastasis. 2010;27:279–293. doi: 10.1007/s10585-010-9326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grim JE, Knoblaugh SE, Guthrie KA, Hagar A, Swanger J, Hespelt J, et al. Fbw7 and p53 Cooperatively Suppress Advanced and Chromosomally Unstable Intestinal Cancer. Mol Cell Biol. 2012;32:2160–2167. doi: 10.1128/MCB.00305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweet-Cordero A, Tseng GC, You H, Douglass M, Huey B, Albertson D, et al. Comparison of gene expression and DNA copy number changes in a murine model of lung cancer. Genes, Chromosomes and Cancer. 2006;45:338–348. doi: 10.1002/gcc.20296. [DOI] [PubMed] [Google Scholar]
- 31.Trobridge P, Knoblaugh S, Washington MK, Munoz NM, Tsuchiya KD, Rojas A, et al. TGF-β Receptor Inactivation and Mutant Kras Induce Intestinal Neoplasms in Mice via a β-Catenin-Independent Pathway. Gastroenterology. 2009;136:1680–1688. e7. doi: 10.1053/j.gastro.2009.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bojovic B, Crowe DL. Dysfunctional telomeres promote genomic instability and metastasis in the absence of telomerase activity in oncogene induced mammary cancer. Mol Carcinog. 2013;52:103–117. doi: 10.1002/mc.21834. [DOI] [PubMed] [Google Scholar]
- 33.Liu ML, Von Lintig FC, Liyanage M, Shibata MA, Jorcyk CL, Ried T, et al. Amplification of Ki-ras and elevation of MAP kinase activity during mammary tumor progression in C3(1)/SV40 Tag transgenic mice. Oncogene. 1998;17:2403–2411. doi: 10.1038/sj.onc.1202456. [DOI] [PubMed] [Google Scholar]
- 34.Standfuss C, Pospisil H, Klein A. SNP microarray analyses reveal copy number alterations and progressive genome reorganization during tumor development in SVT/t driven mice breast cancer. BMC Cancer. 2012;12:380. doi: 10.1186/1471-2407-12-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smolen GA, Muir B, Mohapatra G, Barmettler A, Kim WJ, Rivera MN, et al. Frequent Met Oncogene Amplification in a Brca1/Trp53 Mouse Model of Mammary Tumorigenesis. Cancer Res. 2006;66:3452–3455. doi: 10.1158/0008-5472.CAN-05-4181. [DOI] [PubMed] [Google Scholar]
- 36.Podsypanina K, Li Y, Varmus HE. Evolution of somatic mutations in mammary tumors in transgenic mice is influenced by the inherited genotype. BMC Med. 2004;2:24. doi: 10.1186/1741-7015-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett CN, Green JE. Unlocking the power of cross-species genomic analyses: identification of evolutionarily conserved breast cancer networks and validation of preclinical models. Breast Cancer Research. 2008;10:213. doi: 10.1186/bcr2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chin L, Hahn WC, Getz G, Meyerson M. Making sense of cancer genomic data. Genes & Development. 2011;25:534–555. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding Z, Wu C-J, Jaskelioff M, Ivanova E, Kost-Alimova M, Protopopov A, et al. Telomerase Reactivation following Telomere Dysfunction Yields Murine Prostate Tumors with Bone Metastases. Cell. 2012;148:896–907. doi: 10.1016/j.cell.2012.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hodgson JG. Copy Number Aberrations in Mouse Breast Tumors Reveal Loci and Genes Important in Tumorigenic Receptor Tyrosine Kinase Signaling. Cancer Res. 2005;65:9695–9704. doi: 10.1158/0008-5472.CAN-05-0755. [DOI] [PubMed] [Google Scholar]
- 41.Kim M, Gans JD, Nogueira C, Wang A, Paik J-H, Feng B, et al. Comparative Oncogenomics Identifies NEDD9 as a Melanoma Metastasis Gene. Cell. 2006;125:1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, et al. Identification and Validation of Oncogenes in Liver Cancer Using an Integrative Oncogenomic Approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montagna C, Andrechek ER, Padilla-Nash H, Muller WJ, Ried T. Centrosome abnormalities, recurring deletions of chromosome 4, and genomic amplification of HER2/neu define mouse mammary gland adenocarcinomas induced by mutant HER2/neu. Oncogene. 2002;21:890–898. doi: 10.1038/sj.onc.1205146. [DOI] [PubMed] [Google Scholar]
- 44.Weaver Z, Montagna C, Xu X, Howard T, Gadina M, Brodie SG, et al. Mammary tumors in mice conditionally mutant for Brca1 exhibit gross genomic instability and centrosome amplification yet display a recurring distribution of genomic imbalances that is similar to human breast cancer. Oncogene. 2002;21:5097–5107. doi: 10.1038/sj.onc.1205636. [DOI] [PubMed] [Google Scholar]
- 45.Weaver ZA, McCormack SJ, Liyanage M, du Manoir S, Coleman A, Schrock E, et al. A recurring pattern of chromosomal aberrations in mammary gland tumors of MMTV-cmyc transgenic mice. Genes Chromosomes Cancer. 1999;25:251–260. [PubMed] [Google Scholar]
- 46.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maser RS. Connecting Chromosomes, Crisis, and Cancer. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- 48.Maser RS, Wong KK, Sahin E, Xia H, Naylor M, Hedberg HM, et al. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Mol Cell Biol. 2007;27:2253–2265. doi: 10.1128/MCB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Padilla-Nash HM, McNeil NE, Yi M, Nguyen QT, Hu Y, Wangsa D, et al. Aneuploidy, oncogene amplification and epithelial to mesenchymal transition define spontaneous transformation of murine epithelial cells. Carcinogenesis. 2013;34:1929–1939. doi: 10.1093/carcin/bgt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diede SJ, Yao Z, Keyes CC, Tyler AE, Dey J, Hackett CS, et al. Fundamental differences in promoter CpG island DNA hypermethylation between human cancer and genetically engineered mouse models of cancer. Epigenetics. 2013;8:1254–1260. doi: 10.4161/epi.26486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.