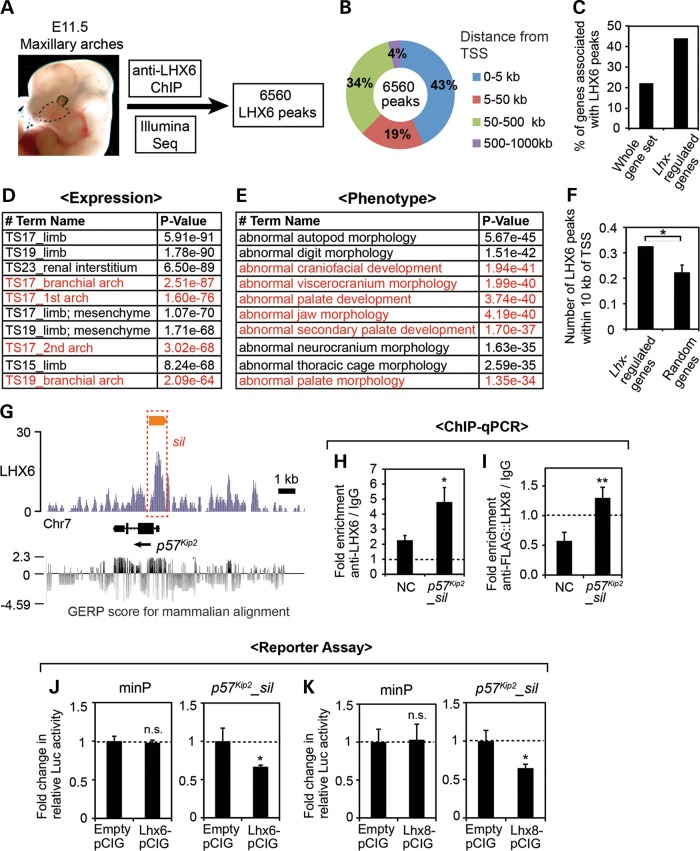
Figure 5.
Identification of an LHX target cis-regulatory element in p57Kip2 locus through LHX6 ChIP-seq. (A) Summary of the ChIP-seq experiment. The dotted line demarcates the maxillary arch. (B) Distribution of the LHX6 peaks relative to the nearest TSS was analyzed using GREAT (37). (C) Comparison of the percentage of the genes associated with LHX6 peaks among the whole-genome gene set, which comprises 20 221 well-annotated genes selected by GREAT, and among the 212 Lhx-regulated genes from our transcriptional profiling. (D and E) Results of the gene ontology analysis of the LHX6 peaks by GREAT. Top ten most enriched terms in the categories of ‘Expression’ and ‘Phenotype’ are listed. The entries that are relevant to craniofacial development are in red. (F) Comparison of the number of LHX6 peaks within 10 kb of the TSS of Lhx-regulated genes and random genes, shown as the average number per gene. The error bar is a 95% confidence interval. *Z score >1.98, or P < 0.05. (G) LHX6 ChIP-seq result at the p57Kip2 locus is visualized by IGB (38). The y-axis corresponds to the fragment density derived from the Illumina sequencing result (see Materials and Methods for details). The orange bar indicates the LHX6 peak determined by MACS peak-finding algorithm (39). The red box (sil) corresponds to p57Kip2_sil used in H–K. Shown at the bottom are the Genomic Evolutionary Rate Profiling (GERP) scores of the region taken from UCSC Genome Browser (40,41). (H and I) ChIP-qPCR from E11.5 maxillary arch cells for p57Kip2_sil and an NC region (NC) located 10 kb away. The error bars are standard deviations from triplicates, and P-values were calculated between the fold enrichments of NC and p57Kip2_sil. *P < 0.05, **P < 0.01. (J and K) Luciferase reporter assay in primary culture of maxillary arch cells from E11.5 embryos. The relative luciferase activity from the minimal promoter (minP) was not significantly different (n.s., P > 0.05) whether co-transfected with an LHX expression vector or empty pCIG as a control. In contrast, the relative luciferase activity from p57Kip2_sil reporter was reduced upon co-transfection of LHX6-pCIG (J) or LHX8-pCIG (K) compared with empty pCIG. The error bars are standard deviations from triplicates. *P < 0.05.