Fig 1. Isolation with Migration model [30].
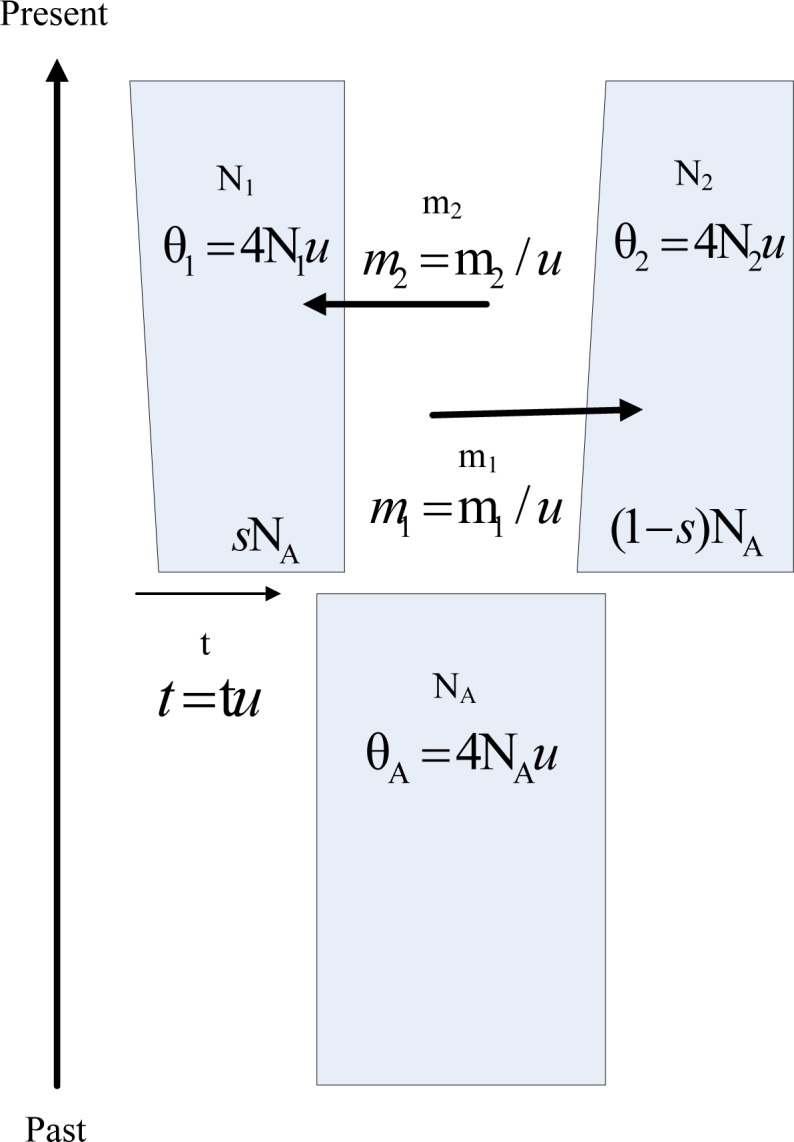
The parameters are effective population sizes (N1, N2, and NA), rates of gene flow (m1 and m2), time of population divergence (t) and proportion of NA that forms the founding population of N1 (s). So the founding sizes of the descendent populations are sNA and (1-s)NA respectively, where 0<s<1. Parameters are all scaled by the neutral mutation rate (u) in IM program, including θ1 = 4N1 u, θ2 = 4N2 u, θA = 4NA u, m 1 = m1 / u, m 2 = m2 / u, t = t / u.
