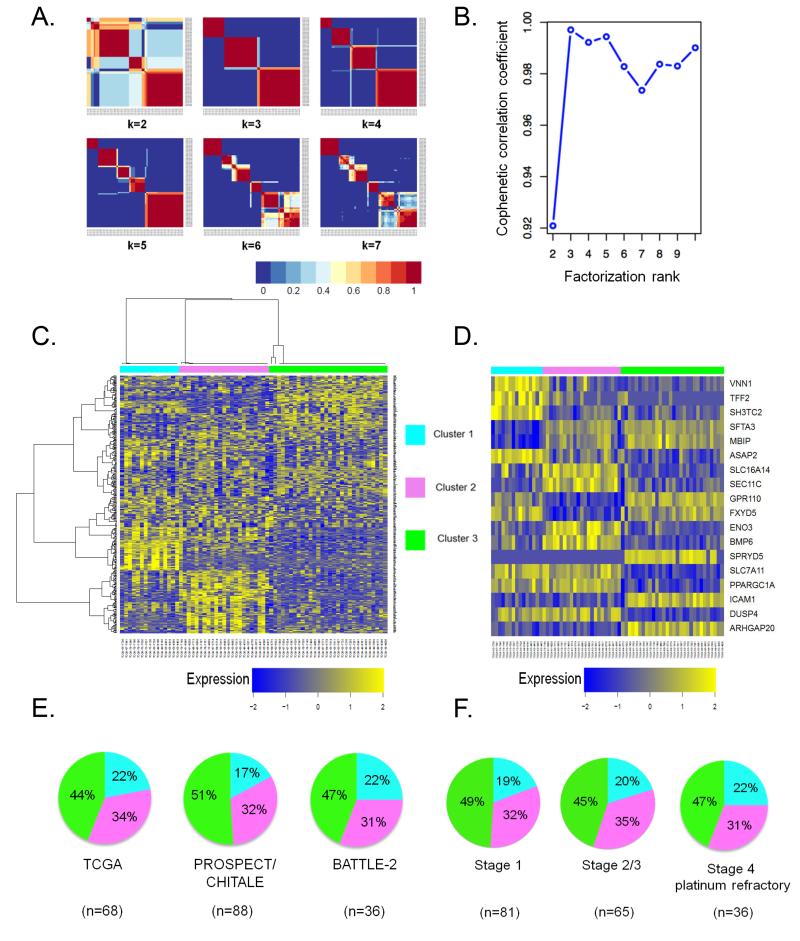
Figure 1. Consensus NMF clustering identifies three robust and reproducible subsets of KRAS-mutant LUAC.
(A) Consensus matrices of 68 KRAS-mutant LUACs from the TCGA dataset, computed for k=2 to k=7.
(B) Cophenetic correlation coefficient plot reveals peak cluster stability for k=3 ranks.
(C) Heatmap depicting mRNA expression levels of 384 genes selected for the NMF algorithm.
(D) Relative expression levels of individual genes that comprise the 18-gene cluster assignment signature in LUACs from the TCGA dataset.
(E) Cluster composition is preserved across distinct datasets of chemotherapy-naïve (PROSPECT, CHITALE) and platinum-refractory (BATTLE-2) KRAS-mutant LUACs.
(F) Subgroup representation is unaffected by increasing disease stage. Stage 4 platinum refractory tumors in this analysis represent the BATTLE-2 clinical cohort.