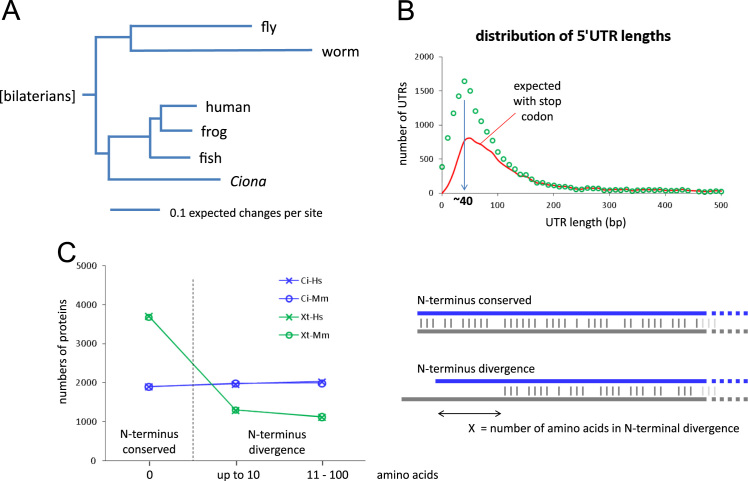
Fig. 1.
Coding genome of Ciona intestinalis. (A). Phylogenetic position of Ciona intestinalis relative to major model organisms, with branch length indicating degree of amino acid divergence (adapted from Putnam et al. (2007)). (B) Length distribution of 5′ UTRs in Ciona intestinalis determined from assembled EST sequence where open reading frame is probably complete. Red line indicates the proportion at any given length expected to include at least one in-frame stop codon. (C) Lack of conservation of N-terminus of Ciona intestinalis proteins relative to well annotated model systems, and compared to Xenopus tropicalis. Comparison of BLASTp alignment data using sets of mutual orthologs between Ciona intestinalis, Xenopus tropicalis, and either human or mouse. Schematic of BLAST alignments indicates how N-terminus divergence is measured.