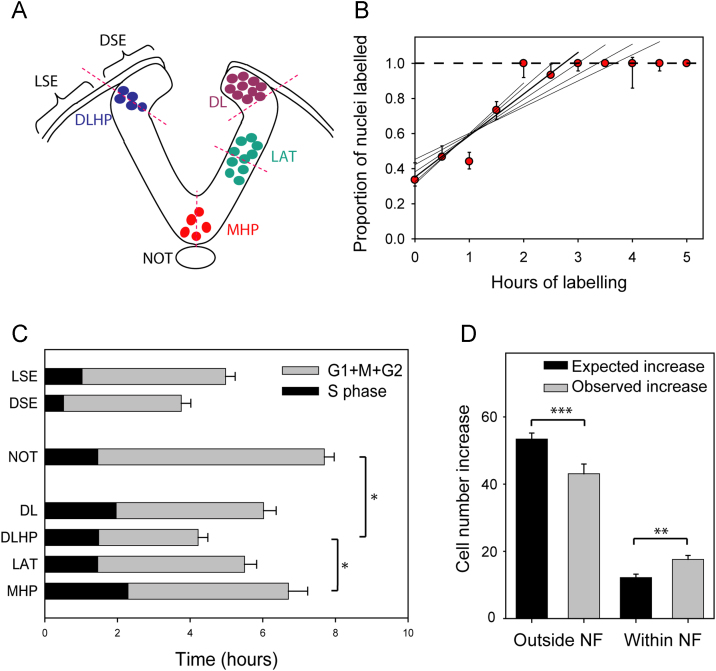
Fig. 6.
Cell cycle analysis during neural plate bending. (A) Diagram of elevated Mode 2 neural plate with attached surface ectoderm, showing tissues included in the analysis: DL, dorsolateral neural plate; DLHP, dorsolateral hinge point; DSE, dorsal surface ectoderm; LAT, lateral non-bending neural plate; LSE, lateral surface ectoderm; MHP, median hinge point; NOT, notochord. (B) Representative data from a 3H-thymidine labelling experiment. An increasing proportion of nuclei are labelled by continuous exposure to 3H-thymidine until, in this example, 100% of nuclei are labelled after 2 h. Regression analysis yields a family of lines, each generated by including one additional post-2 h time point. The best-fit regression line (bold) was chosen to calculate cell cycle length in each data set. See also Fig. S3. (C) Total cell cycle length (hours+SEM) as calculated for tissues in (A). Calculated lengths of S phase (black) and combined G1+M+G2 (grey) are shown. Total cell cycle length varies significantly between tissues (p<0.001; 1-way ANOVA), with DLHP exhibiting a significantly shorter cell cycle length than either MHP or notochord (*p<0.05; post-hoc Bonferroni t-tests). (D) Cell number increase in neural plate between flat and elevated stages of PNP closure (data from Table 1). Expected increase (black bars) is based on cell cycle length and observed increase (grey bars) is based on cell number counts. Expected increase significantly exceeds observed for neural plate outside the neural fold (NF)(***p=0.02; t-test), whereas expected increase is significantly less than observed for neural plate within the NF (**p=0.01).