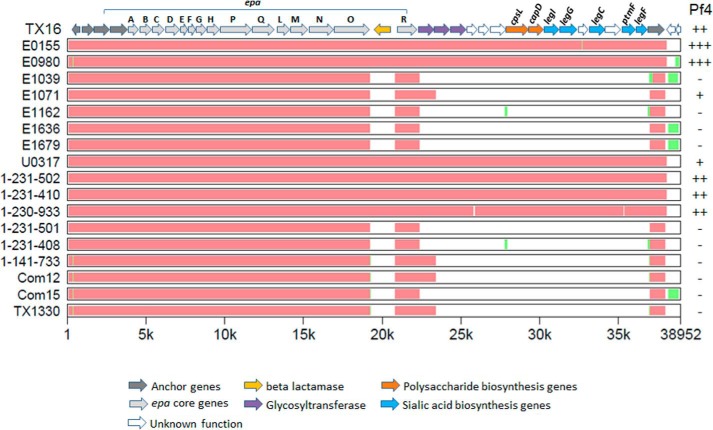
FIGURE 8.
Conservation of Tx16 legionaminic acid biosynthesis gene cluster (legIGCF) among E. faecium strains. Orthologous sequences of E. faecalis epa, polysaccharide biosynthesis, and sialic acid biosynthesis genes (including anchor genes at two ends) were selected from each E. faecium genome and aligned by MultiPipMaker using default settings (38). As the reference, Tx16 genes are shown at top, with individual genes represented by arrows. Genes with homology to S. agalactiae and S. aureus CPS biosynthesis genes are labeled cpsL and capD, respectively. Orthologues of C. jejuni legionaminic acid biosynthesis genes are labeled accordingly (supplemental Table S2) (36). The x axis denotes the location of evolutionarily conserved regions in relation to the Tx16 sequence (22). Red regions represent areas of high sequence conservation (at least 100 bp with no gaps and >70% identity), and green represents regions where an alignment could be made but with lower sequence conservation. White regions denote the absence of gene sequences present in Tx16. The reactivity of strains with antisera to legionaminic acid heteroglycan Pf4 is indicated (see Table 5 for values and supplemental Table S1 for key). GenBankTM accession numbers for genome sequences are as follows: Tx16 (NC_017960); E0155 (AUWX00000000); E0980 (ABQA01000000); E1039(ACOS01000000); E1071 (ABQI01000000); E1162 (ABQJ01000000); E1636 (ABRY01000000); E1679 (ABSC01000000); U0317 (ABSW01000000); 1-231-502 (ACAX01000000); 1-231-410 (ACBA01000000); 1-230-933 (ACAS01000000); 1-231-501 (ACAY01000000); 1-231-408 (ACBB01000000); 1-141-733 (ACAZ01000000); Com12 (ACBC01000000); Com15 (ACBD01000000); and TX1330 (ACHL01000000).