FIGURE 4.
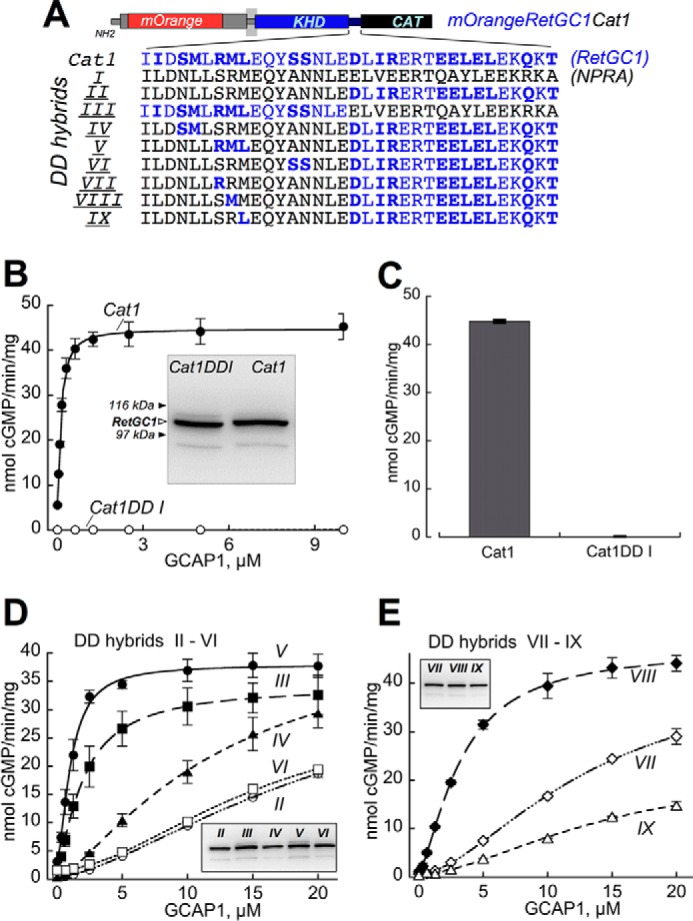
Sensitivity of RetGC1/NPRA dimerization domain hybrids to GCAP-dependent activation. A, portions of the dimerization domain coded by NPRA gene (black) in the Cat1DD I chimera were replaced by those coded by RetGC1 gene (blue). The RetGC1-specific amino acid residues are highlighted in bold (note that some residues coded by the two respective different genes are identical). CAT, catalytic domain. B and C, GCAPs activate Cat1 but fail to activate Cat1DD I. B, dose dependence of Cat1 (●) versus Cat1DD I (○) activation by GCAP1 at ≪10 nm free [Ca2+] and 6 mm free [Mg2+]. Inset, immunoblotting of the two chimeric constructs expressed in HEK293 membranes probed with anti-RetGC1 KHD antibody (37); the filled arrows indicate positions of β-galactosidase (116 kDa) and phosphorylase b (97 kDa). C, the membranes expressing Cat1 and Cat1DD I were reconstituted with 10 μm GCAP2 at ≪10 nm free [Ca2+] and 6 mm [Mg2+]. D and E, dose dependence of GCAP1 activation of the Cat1 chimeras containing NPRA/RetGC1 hybrids II–VI (D) and VII–IX (E) depicted in A. Insets, immunoblotting of 7% SDS-polyacrylamide gel loaded with 10-μl aliquots of the respective membrane fractions (molecular weight markers are not shown because of space constraints). The activities in assays shown in B–E were standardized by the protein content. The data were fitted using Synergy KaleidaGraph 4 utilizing the standard Levenberg-Marquardt algorithm of nonlinear least square routines assuming a Hill function: a = (amax − amin)/(1 + ([GCAP]/(K1/2GCAP)h) + amin where a is the activity of RetGC in the assay; amin and amax are the minimal and the maximal activities, respectively; [GCAP] is the concentration of GCAP, K1/2GCAP is the GCAP concentration required for half-maximal activation, and h is the Hill coefficient. The K1/2GCAP was low for DD V (1.1 ± 0.06 μm) and DD III (2 ± 0.03 μm) but sharply increased in DD hybrids II, IV, and VI (21.5 ± 0.70, 12.5 ± 1.2, and 15 ± 1.8, respectively). The K1/2GCAP values for DD VII, VIII, and IX hybrids in E were 15 ± 2.3, 3.2 ± 0.16, and 22 ± 12 μm, respectively (the low activity in DD hybrid IX increased the error for the K1/2GCAP extraction from the fitting). The assay conditions are described under “Experimental Procedures.” Error bars are S.E.
