Figure 1.
Single-Cell Transcriptional Profiling Captures Molecular Spaces of Lineage Commitment
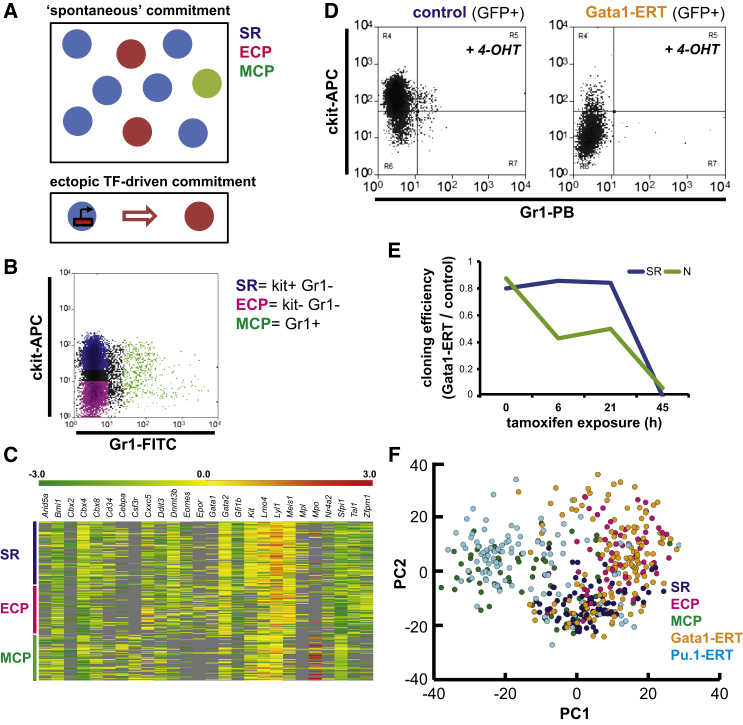
(A) Depiction shows the two modes of commitment surveyed in this study: (top) hierarchically related SR cells, ECPs, and MCPs in equilibrium in a multipotent cell culture system; (bottom) unilineage commitment of SR cells driven by a single TF.
(B) Flow cytometry plot shows co-existing SR, ECP, and MCP cells in an FDCPmix culture under SR conditions.
(C) Heatmap of expression profiles of 26 genes in individual FDCPmix SR, ECP, and MCP cells. The gene panel represents the consensus analyzed in sufficient cell numbers in all compartments between replicate experiments. Data are Z score-normalized ΔCt values; undetectable expression, gray.
(D) Flow cytometry plots show FDCPmix SR cells transduced with control empty vector or with a GATA1-ERT fusion after 21-hr activation with tamoxifen (4-OHT, 1 μM).
(E) Cloning efficiency of control and Gata1-ERT GFP+ cells cultured under SR and neutrophil differentiation (N) conditions during a time course of 4-OHT activation. Sampling times correspond to those of single-cell qRT-PCR analysis. At each time point, 60 individual cells of each genotype were plated into SR or N conditions, and individually seeded wells were inspected at regular intervals for a 7-day period.
(F) PCA plot of the transcriptional profiles of individual FDCPmix cells undergoing distinct modes of lineage commitment. The first two PC explain 31% of the data variance; n = 82 (SR), 60 (ECP), 59 (MCP), 147 (Gata1-ERT), and 103 (Pu.1-ERT). A consensus set of 22 genes was analyzed in all five compartments.