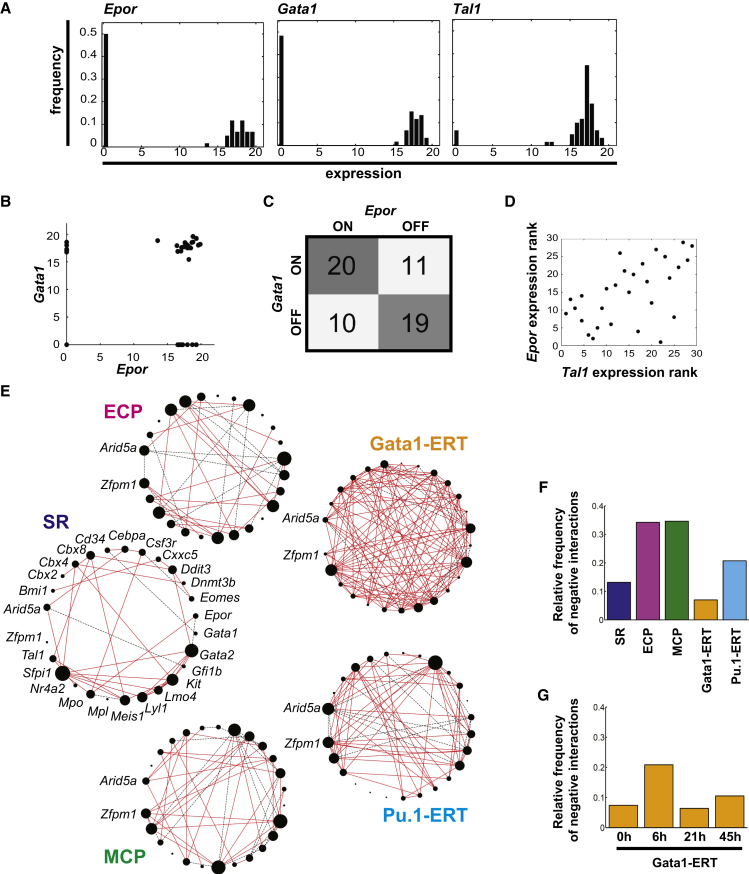
Figure 2.
Combined Single-Cell Transcriptional Network Inference Methods Implicate Ddit3 in Lineage Specification
(A) Representative gene expression distributions for Epor, Gata1, and Tal1 in ECPs are shown.
(B) Scatterplot of Gata1 and Epor single-cell expression highlights the dual aspect of the data with both binary (on/off) and continuous (expression-level) components.
(C) Contingency table summarizing on/off combination patterns of individual cells for Epor and Gata1. OR quantifies the diagonal versus off-diagonal of this matrix to infer significant positive and negative associations in the binary component of the data. Gata1 and Epor show significant positive association (OR = 3.18; lower95CI > 1).
(D) Scatterplot of Epor and Tal1 expression ranks in co-expressing cells. Epor and Tal1 show significant positive correlation in the continuous component of the data inferred by Spearman rank correlation (r = 0.56; p = 0.002).
(E) Single-cell transcriptional networks in SR, ECP, MCP, Gata1-ERT, and Pu.1-ERT compartments were inferred by combined use of OR and Spearman rank correlation. Solid red lines, positive associations; dashed black lines, negative associations. Node size is proportional to the relative connectivity in each network.
(F) Proportion of negative interactions in the networks in (E) is shown.
(G) Proportion of negative interactions in Gata1-ERT networks at each time point is shown.