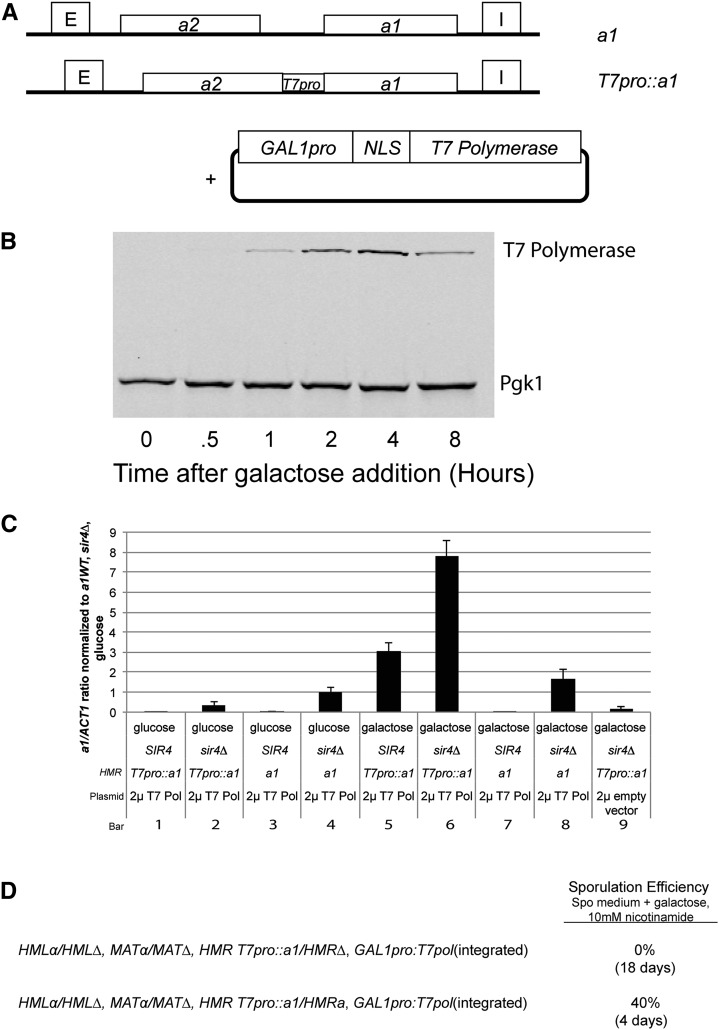
Figure 2.
Quantifying transcription and translation of a1 transcripts from HMR T7pro::a1. (A) A schematic of T7pro::a1 at HMR in comparison to wild-type HMRa1. Twenty bp of the T7 minimal optimum promoter (Ujvári and Martin 1997) replaced the region between a2 and 5 bp upstream of the initiation codon for a1 at HMR. The schematic of the 2 μ plasmid (pJR3207) carrying the nuclear localization signal–enhanced T7 RNA polymerase gene is also shown. (B) Protein immunoblot of T7 RNA polymerase protein levels before and upon galactose induction in CSM-leu medium. Pgk1 levels served as a loading control. (C) Quantitation of a1 transcripts as determined by qRT-PCR of wild-type a1 and T7pro::a1 at HMR in SIR4 and sir4∆, with or without T7 RNA polymerase. All a1 expression values were normalized to ACT1 mRNA from the same sample, and that ratio was further normalized to the a1/ACT1 mRNA ratio in HMRa1 sir4∆ strains grown in glucose. All cultures were seeded from saturated overnight growth in the indicated media. cDNA synthesis was primed using random hexamers. Each bar represents the average and SE of three biological replicates. (D) Diagram of strain genotypes used to test possible translation of a1 transcripts made by T7 RNA polymerase. The top line shows a diploid whose only source of a1 mRNA would be transcribed from T7pro::a1 by T7 RNA polymerase. The bottom line shows an isogenic control diploid with a source of wild-type a1 mRNA at HMR. Ten mM nicotinamide was used to derepress HML and HMR and allow for expression of mating-type information normally silenced by Sir proteins. Sporulation requires both a1 and α2 proteins and the formation of a heterodimer to proceed. Sporulation efficiency was measured as a percentage of cells that formed tetrads.