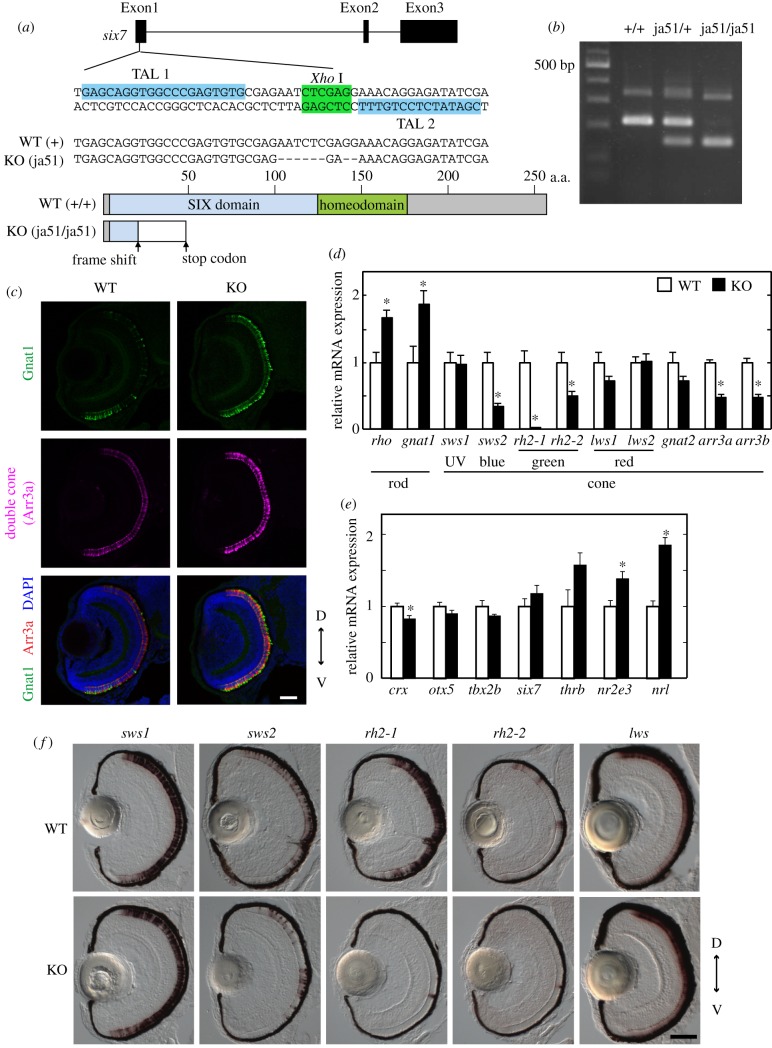
Figure 3.
Analysis of the WT zebrafish and the six7 KO zebrafish at the larval stage. (a) Top: exon–intron organization and partial nucleotide sequences of zebrafish six7. The binding sites of the left and right TALENs are highlighted in blue. The recognition site of the endonuclease XhoI is highlighted in green. Middle: the nucleotide sequence of the KO (ja51 mutant) fish is compared with the WT sequence. Deletions are indicated by dashes. Bottom: schematic of Six7 and its mutant protein. (b) Genotyping of the six7 KO zebrafish. See details in the electronic supplementary material, methods. (c) Immunofluorescence of ocular sections of the WT and KO larvae at 3.5 dpf. The ocular sections were immunostained with the anti-Gnat1 antibody for rods (green) and with the zpr1 antibody for double cones (i.e. red and green cones; magenta). DAPI staining identified all the cell nuclei (blue). Scale bar, 40 µm. (d,e) Expression profiles of the phototransduction component genes (d) and the transcription factors contributing to the photoreceptor development in mouse and/or zebrafish (e) in the WT and six7 KO larval eyes at 3.5 dpf. The mRNA levels were quantified by RT-qPCR. The data were represented by mean + s.e.m. (n = 5). Statistical significance between WT and six7 KO data is shown by the asterisks (*p < 0.05 by Student's t-test). (f) Expression profiles of the cone opsin genes by in situ hybridization using the larval eyes of the WT and KO at 3.5 dpf. The lws probe was designed to recognize both of the lws1 and lws2 opsin genes. D, dorsal side; V, ventral side. Scale bar, 50 µm. (Online version in colour.)