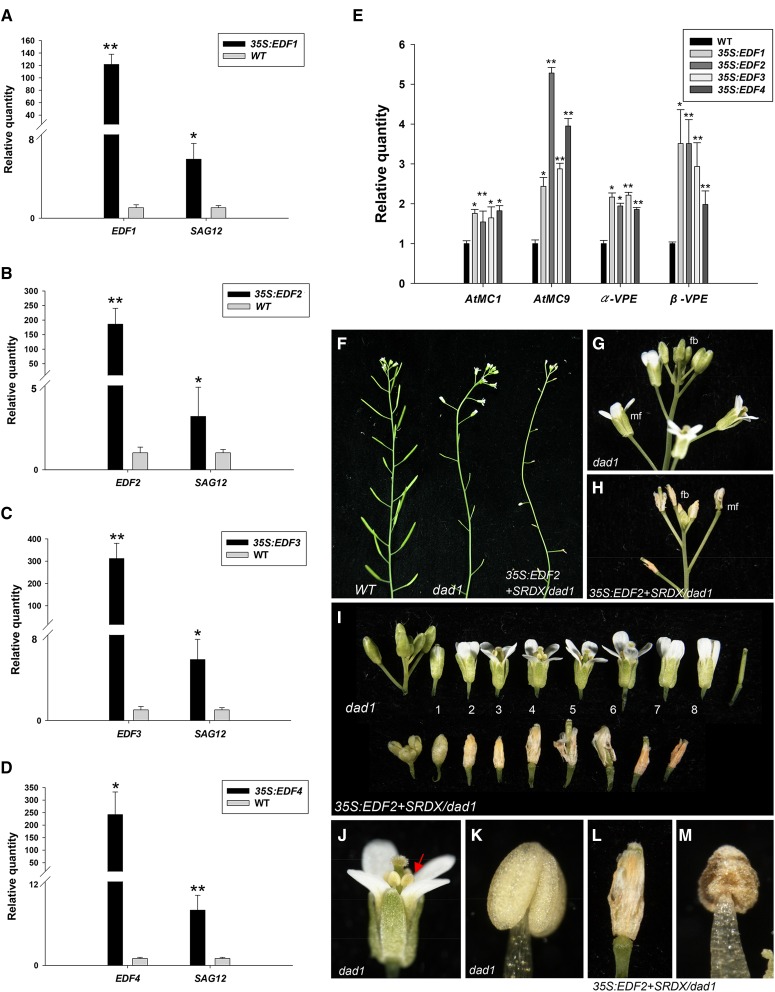
Figure 2.
The detection of senescence-associated gene expression in 35S:EDF plants and a phenotypic analysis of flower senescence and abscission in dad1 plants ectopically expressing the EDF2+SRDX gene. A to D, The expression of the SAG12 gene in 35S:EDF1 (A), 35S:EDF2 (B), 35S:EDF3 (C), and 35S:EDF4 (D) plants. E, The expression of AtMC1, AtMC9, α-VPE, and β-VPE genes in 35S:EDF plants. The mRNA levels were determined by real-time quantitative PCR. Total RNA was isolated from the floral buds and the open flowers at positions 1 to 3 from one wild-type (WT) Col-0 plant and one 35S:EDFs plant. The transcript levels of these genes were determined using two to three replicates and normalized using UBQ10. The expression of each gene is given relative to that of the wild-type plant, which was set at one. The asterisks in A to E indicate a significant difference from the wild-type value. *, P ≤ 0.05; **, P ≤ 0.01. F, The inflorescences of wild-type (left), dad1 (center), and 35S:EDF2+SRDX/dad1 (right) plants. No silique elongation was observed in dad1 and 35S:EDF2+SRDX/dad1 plants. G and H, Close-up of the flower buds (fbs) and mature flowers (mfs) for dad1 (G) and 35S:EDF2+SRDX/dad1 (H) plants from F. I, The flowers along an inflorescence of the dad1 (upper) and 35S:EDF2+SRDX/dad1 (lower) plants. The perianth organs did not senesce and abscised after position 8 for dad1 mutants. The numbers indicate the positions of the flowers. The perianth organs of the flowers in 35S:EDF2+SRDX/dad1 plants were completely senescent and abscised at positions 2 and 3. J and K, Close-up view of a mature flower at position 4 (J) and an indehiscent anther (K) for the dad1 plant from I. An arrow indicates the indehiscent anthers in J. L and M, Close-up view of a mature flower at position 4 (L) and an indehiscent anther (M) of the 35S:EDF2+SRDX/dad1 plant from I. Expression statistical analysis was measured by Student’s t test (each n = 3).