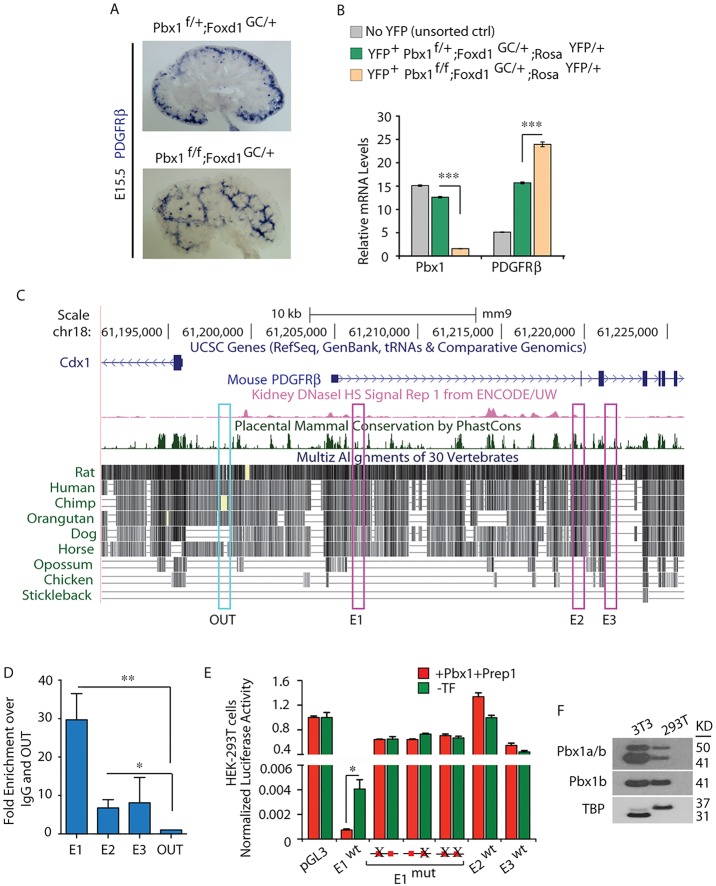
Fig. 5.
Pbx1 homeoprotein regulates Pdgfrb by binding to a cis-regulatory element within the Pdgfrb locus. (A) ISH of Pdgfrb on E15.5 control Pbx1f/+;Foxd1GC/+ and mutant Pbx1f/f;Foxd1GC/+ kidney sections displaying ectopic Pdgfrb mRNA distribution in the mutant kidneys. (B) Loss of Pbx1 in the Foxd1 lineage results in upregulation of Pdgfrb transcript levels. Quantitative RT-PCR analysis of Pdgfrb and Pbx1 mRNAs in FACS-sorted YFP+ cells obtained from E15.5 Pbx1f/+;Foxd1GC/+;RosaYFP/+ and Pbx1f/f;Foxd1GC/+;RosaYFP/+ and cells from control YFP− kidneys. Values were normalized to the housekeeping gene Tbp. Kidney cells were pooled from at least five embryos per genotype. The experiment was performed in technical triplicates. Error bars represent s.e.m.; ***P<0.0001. (C) 5′ portion of the mouse Pdgfrb locus (UCSC Genome Browser, mm9). Conservation plot across vertebrate species; dark green peaks indicate highest conservation across mammals; pink peaks indicate sites of open chromatin by the presence of DNaseI hypersensitivity; pink boxes (E1, E2, E3) highlight conserved non-coding regions containing consensus Pbx-Meis/Prep binding sequences. (D) ChIP-qPCR of Pbx binding to two Pdgfrb cis-regulatory regions in E15.5 wild-type kidneys. Fold enrichment over IgG relative to amplification within a non-conserved region (OUT) is shown. Error bars represent s.e.m. from six IPs with an anti-PanPbx antibody from two independent chromatin preparations. Significant binding of Pbx to E1 (**P=0.0018) and E2 (*P=0.025) was observed. (E) Pbx-dependent transcription reporter assays in HEK-293T cells transfected with the indicated plasmids. Representative experiment (out of eight) is shown. Data were calculated as firefly to Renilla luciferase ratios of each reporter construct, normalized to empty TK-pGL3 firefly to Renilla ratios (set to 1). Wild-type (wt) E1 strongly repressed firefly luciferase expression, which is significantly enhanced upon Pbx1b and Prep1 overexpression (cumulative *P=0.011). Disruption of one or both Pbx-Meis/Prep binding motifs in E1 abrogated transcriptional repressive activity. Activities of E2 and E3 were negligible. (F) Immunoblot revealing endogenous Pbx1a and Pbx1b proteins in 293T and 3T3 cell lines. TBP, loading control. See also supplementary material Fig. S4.