Fig 2. Electrophoretic mobility shift assays with probe Sp1-1+2 (A) and probe Sp1-3 (B).
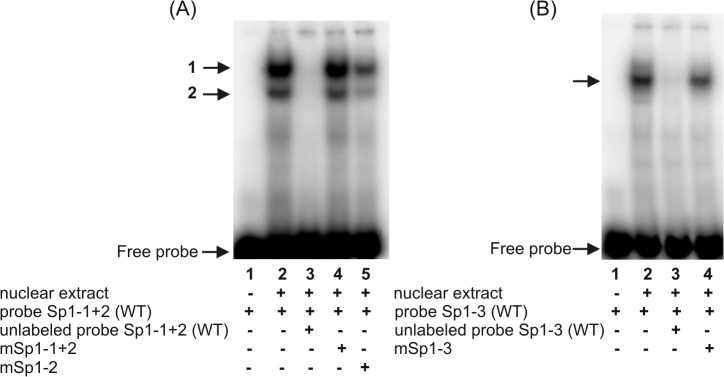
Probe Sp1-1+2 − double-stranded oligonucleotide representing the -646/-611 sequence of the hFCGRT promoter, containing the potential transcriptional regulatory Sp1 motifs at positions -641 and -635; probe Sp1-3 − ds-oligonucleotide corresponding to the sequence -320/-293 of the hFCGRT promoter containing a putative Sp1 binding site at position -313. Probes were end-labeled with [γ32P]ATP] and incubated with nuclear extract (12 μg) in the absence of competitor (A and B, lanes 2) or in the presence of a 100-fold molar excess competitor: Sp1-1+2(WT)–unlabeled wild-type probe Sp1-1+2 (A, lane 3), Sp1-3(WT)–unlabeled wild-type probe Sp1-3 (B, lane 3), mSp1-1+2 – unlabeled probeSp1-1+2 containing mutation in the Sp1 sequence at positions -641 and -635 (A, lane 4), mSp1-2 – unlabeled probe Sp1-1+2 containing mutation in the Sp1 sequence at position -635 (A, lane 5), mSp1-3 – unlabeled probe Sp1-3 containing mutation in the Sp1 sequence at position -313 (B, lane 4). Labeled probe Sp1-1+2(WT) alone (A, lane 1), labeled probe Sp1-3(WT) alone (B, lane 1). DNA-protein complexes were resolved on 5% non-denaturing polyacrylamide gels and analyzed in a phosphor imager (Typhoon 8600) using ImageQuant software (Molecular Dynamics). Positions of specific DNA-protein complexes are indicated by arrows.
