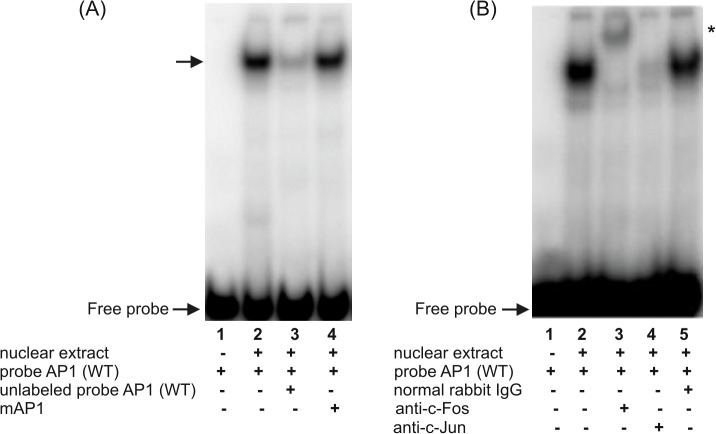
Fig 4. Characterization of the putative AP-1 binding site in the hFCGRT promoter by EMSA (A) and supershift analysis (B).
EMSA and supershift experiments were performed using the AP1 probe corresponding to the hFCGRT promoter sequence between the nucleotides -284 and -254, which contained the putative AP-1 binding site at position -276. The arrow indicates the specific complex formed during incubation of the 32P-labeled wild-type AP1 probe, AP1(WT), with the nuclear extract from differentiated THP-1 cells (A, lane 2). Competition experiments were performed in the presence of a 100-fold molar excess of unlabeled probe AP1(WT) (A, lane 3) or in the presence of its mutated version, in which the putative AP-1 binding site at position -276 was mutagenized–mAP1 (A, lane 4). Supershift experiments were performed by preincubating the nuclear extract with the rabbit polyclonal anti-c-Fos antibody (B, lane 3), anti-c-Jun antibody (B, lane 4), normal rabbit IgG (B, lane 5), prior to the addition of 32P-labeled wild-type AP1 probe–AP1(WT). Labeled probe AP1(WT) alone (A and B, lanes 1); labeled probe AP1(WT) incubated with nuclear extract in the absence of antibodies (B, lane 2). Position of the shifted complex is marked by an asterisk. Results were analyzed by a phosphor imager.