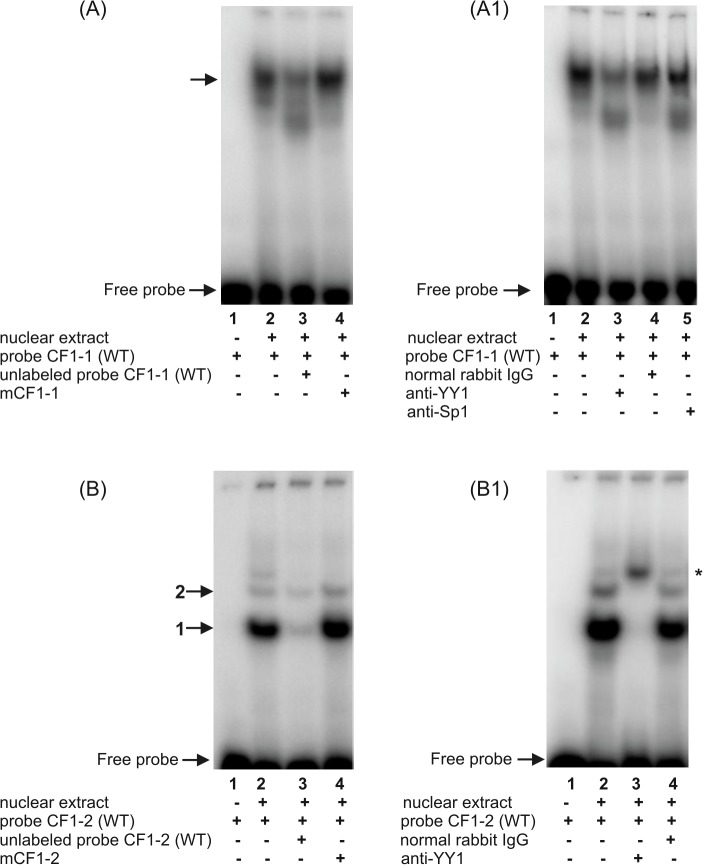
Fig 5. Characterization of two potential CF1/YY1 regulatory motifs in the -660/-233 fragment of the hFCGRT promoter by EMSA (A, B), and supershift analysis (A1, B1).
EMSA and supershift experiments were carried out using the following probes: CF1-1 − double-stranded oligonucleotide representing the hFCGRT promoter sequence between the nucleotides -593 to -565, containing the putative CF1/YY1 binding site at position -586 (A, A1); CF1-2 – ds-oligonucleotide corresponding to the -362/-337 sequence of the hFCGRT promoter containing the potential CF1/YY1 transcriptional regulatory motif at position -357 (B, B1). In EMSA, 32P-labeled wild-type probes were incubated with nuclear extract in the absence of competitor (A and B, lanes 2) or in the presence of a 100-fold molar excess of competitor: CF1-1(WT)–unlabeled wild type probe CF1-1 (A, lane 3), CF1-2 (WT)–unlabeled wild type probe CF1-2 (B, lane 3), mCF1-1 – probe CF1-1 with mutation in the CF1 element at position -586 (A, lane 4), mCF1-2 – probe CF1-2 containing mutation in the CF1 site at position -357 (B, lane 4). Supershift experiments were performed by preincubating the nuclear extract with the anti-YY1 rabbit polyclonal antibody (A1 and B1, lanes 3), anti-Sp1 antibody (A1, lane 5), normal rabbit IgG (A1 and B1, lanes 4), prior to the addition of 32P-labeled wild-type probe: CF1-1(WT) (A1); CF1-2(WT) (B1). Labeled probes incubated with nuclear extract in the absence of antibodies (A1 and B1, lanes 2). Labeled probe CF1-1(WT) alone (A and A1, lanes 1); labeled probe CF1-2 (WT) alone (B and B1, lanes 1). Arrows indicate the specific DNA-protein complexes and the asterisk indicates the supershift complex. Results were analyzed by a phosphor imager.