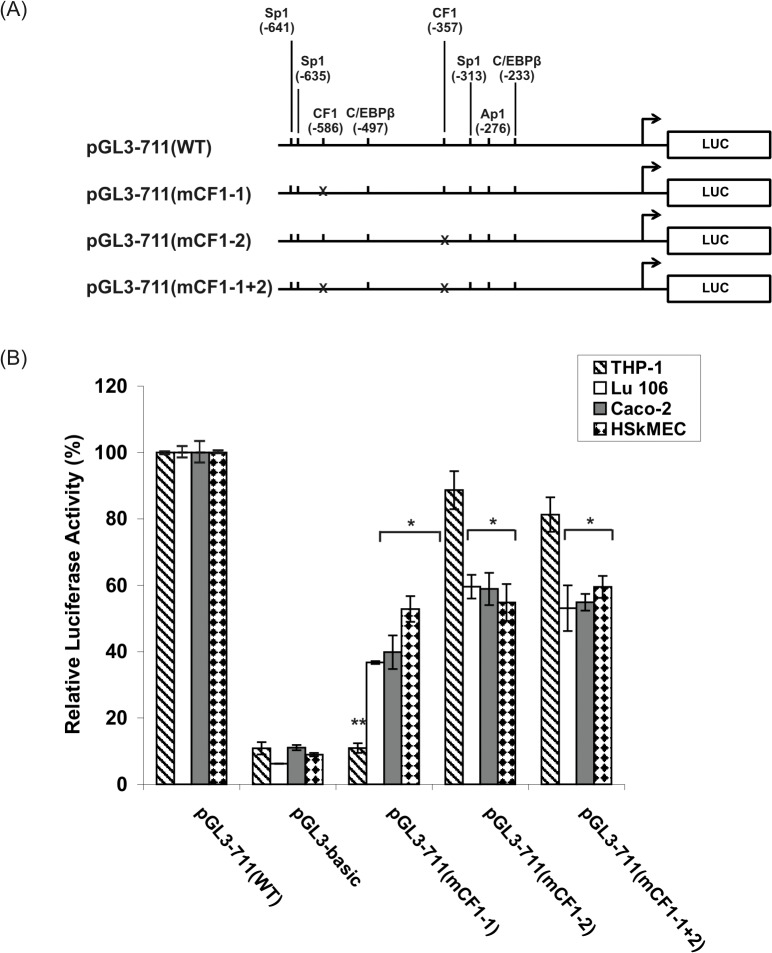
Fig 10. Functional analysis of the CF1 binding sites within the hFCGRT promoter.
(A) Schematic diagram representing the wild-type promoter construct and the mutant promoter constructs. Mutations are marked with crosses. (B) Summarized results of luciferase activity tests. The mutant constructs pGL3-711(mCF1-1), pGL3-711(mCF1-2), and pGL3-711(mCF1-1+2) are derivatives of the wild-type pGL3-711(WT) with mutation in the CF1 site at positions-586, -357, -586 plus -357, respectively. Transfection and normalization were performed as described in the legend to Fig 8. All results represent means ± SD of three to six independent experiments performed in duplicate and plotted as a percentage of the normalized activity of the wild-type pGL3-711(WT) construct, which is defined as 100%. The promoter activity of the labeled construct is significantly decreased compared to the wild-type construct; *P < 0.01 vs. pGL3-711(WT); **P < 0.01 compared with pGL3-711(mCF1-2) and pGL3-711(mCF1-1+2) in THP-1.