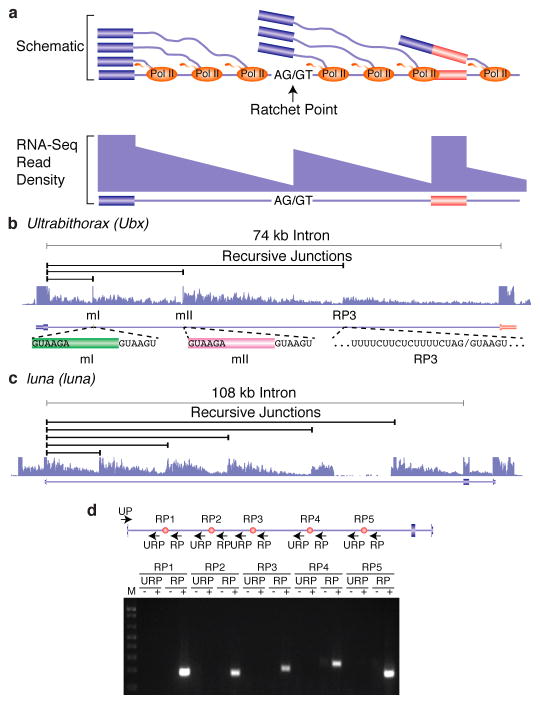
Figure 1. Identification and validation of recursive splice sites in Drosophila.
a, Schematic diagram of nascent pre-mRNA transcripts during co-transcriptional splicing and the corresponding read density that would be observed in total RNA-Seq data. Note the sawtooth pattern created by the 5′ to 3′ gradient of RNA-Seq read density from the exon to the downstream ratchet point and splice site. b, Example of total RNA-Seq data for the Ubx gene which is known to contain three recursive splice sites. Also shown are the splice junction reads supporting recursive splicing at each site. c, Example of five recursive splice sites identified in luna. Shown are the recursive junctions identified and the overall RNA-Seq read density from all samples (blue). d, RT-PCR validation of the luna ratchet points (red dots) using primers in the upstream constitutive exon and flanking the putative ratchet points (UP). The RP primers are expected to yield RT-PCR products if the constitutive exon is spliced to the ratchet point. The URP primers, which are upstream of each ratchet point, serve as negative controls.