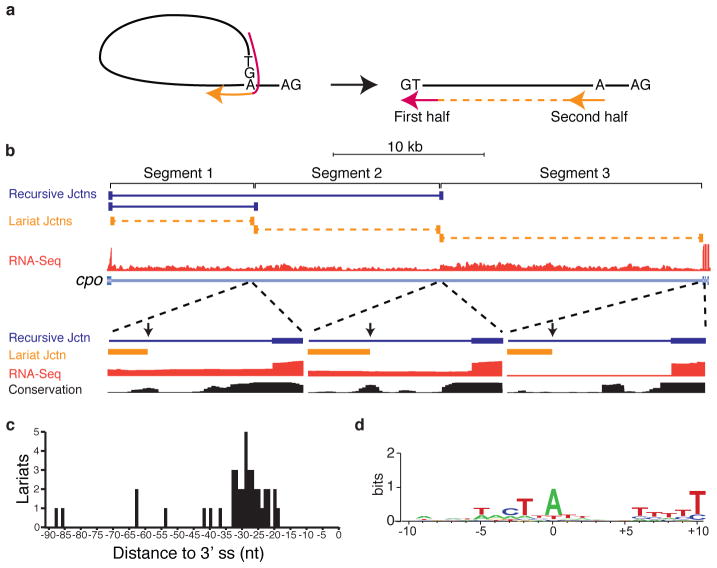
Figure 2. Identification of recursive lariat introns in Drosophila.
a, RNA-Seq reads (red and orange indicate the first and second half of an individual read) that traverse a 5′ splice site-branchpoint junction would align to the linear intron as out-of order split-reads. b, Example of recursive lariat introns in cpo. Shown are the recursive junctions identified (blue), the lariat junction reads (orange), and the overall RNA-Seq read density from all samples (red). A magnification of each branch point region is also shown along with the conservation among 16 insects. The positions of the branch points are indicated by the vertical arrows. c, Distribution of the distance of the recursive lariat intron branch points from the 3′ splice sites. d, Sequence logo of the recursive lariat intron branch point sequences.