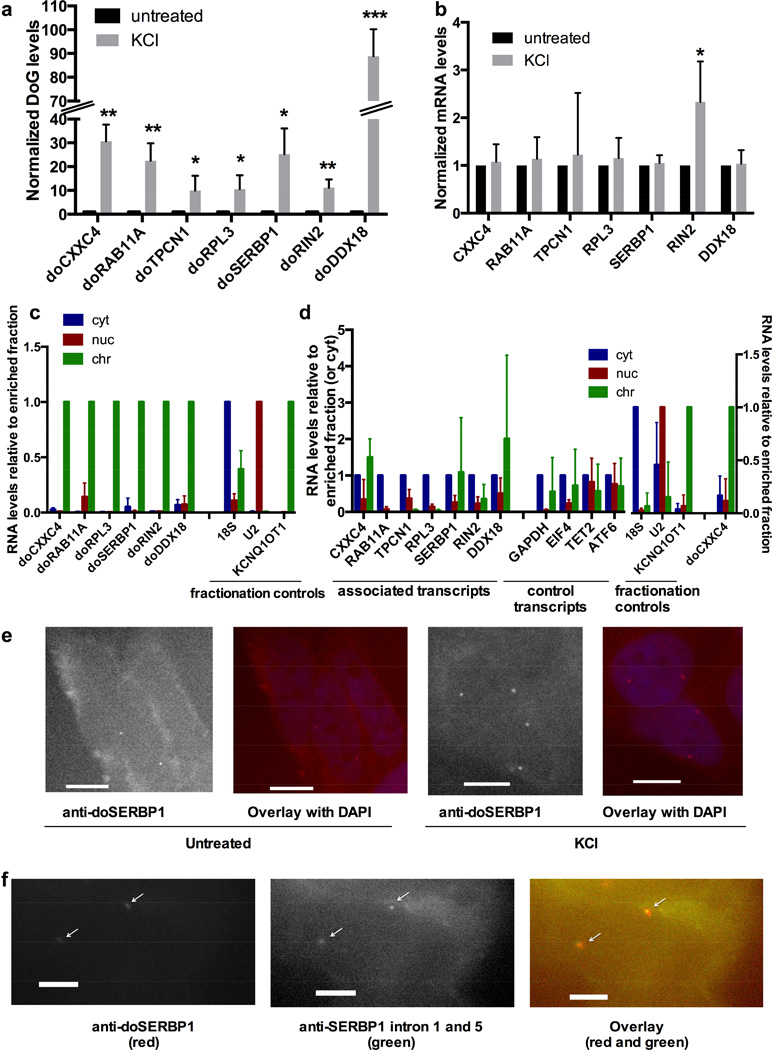
Figure 3. Characterization of DoGs.
a) Validation of DoG induction by KCl (80 mM, 1 h), n=3, except for doTPCN1, where n=4. doCXXC4 from Figure 1a is included for comparison. Figures of pooled data show mean and SD, p-values are denoted as in Figure 1. RNA levels are normalized to GAPDH mRNA in a and b. b) mRNA transcribed from DoG-associated genes are not induced by KCl with the exception of RIN2, which is induced 2-fold, while doRIN2 is induced 10-fold (Figure 3a), n=3, except for RIN2, where n=4. c) qRT-PCR quantitation of DoGs in RNA purified from cytoplasmic, soluble nuclear, or the chromatin fraction. KCNQ1OT1 lncRNA, U2 snRNA and 18S rRNA served as markers for the chromatin (chr), nuclear (nuc), and cytoplasmic (cyt) fraction, respectively, n=3. The enriched fraction is set to 1. All DoGs are significantly enriched in chromatin. d) As in c for DoG-associated mRNAs and control mRNAs, none of which are enriched in chromatin. Due to lack of material, 5 biological replicates were used to reach an n=3 for each target. Fractionation controls were significantly enriched in their respective fractions in both triplicate combinations used, but for simplicity, controls are shown as the mean and SD of all 5 replicates. doCXXC4 is an additional control to verify DoG localization to chromatin. doCXXC4, fractionation controls, RAB11A, RIN2, RPL3, and TPCN1 are significantly enriched in the fraction set to 1 compared to either of the other fractions. Other targets that showed no statistically significant enrichment in any fraction are shown normalized to the cytoplasmic fraction. e) Representative images of FISH using Stellaris probes (Biosearch) for doSERBP1 in untreated and KCl-treated SK-N-BE(2)C cells, confirming KCl-mediated induction and nuclear localization of doSERBP1. Images are at 40x magnification and scale bars indicate 10 µM. f) Representative images of co-staining with FISH Stellaris probes (Biosearch) for intron 1 and 5 of SERBP1 in KCl-treated cells demonstrating that doSERBP1 remains at the site of transcription. Images are at 100x magnification and scale bars indicate 5 µM. See Extended Experimental Procedures for details on FISH probes. See also Figures S3 and S4.