Abstract
Uropathogenic Escherichia coli (UPEC) are phenotypically and genotypically very diverse. This diversity makes it challenging to understand the evolution of UPEC adaptations responsible for causing urinary tract infections (UTI). To gain insight into the relationship between evolutionary divergence and adaptive paths to uropathogenicity, we sequenced at deep coverage (190×) the genomes of 19 E. coli strains from urinary tract infection patients from the same geographic area. Our sample consisted of 14 UPEC isolates and 5 non-UTI-causing (commensal) rectal E. coli isolates. After identifying strain variants using de novo assembly-based methods, we clustered the strains based on pairwise sequence differences using a neighbor-joining algorithm. We examined evolutionary signals on the whole-genome phylogeny and contrasted these signals with those found on gene trees constructed based on specific uropathogenic virulence factors.
The whole-genome phylogeny showed that the divergence between UPEC and commensal E. coli strains without known UPEC virulence factors happened over 32 million generations ago. Pairwise diversity between any two strains was also high, suggesting multiple genetic origins of uropathogenic strains in a small geographic region. Constrasting the whole-genome phylogeny with three gene trees constructed from common uropathogenic virulence factors, we detected no selective advantage of these virulence genes over other genomic regions. These results suggest that UPEC acquired uropathogenicity long time ago and used it opportunistically to cause extraintestinal infections.
Keywords: uropathogen, next-generation sequencing, phylogeny
Introduction
Extraintestinal Escherichia coli (E. coli) are capable of causing various infections, including urinary tract infection (UTI) and meningitis (Foxman, 2002). Approximately 90% of all UTI cases are caused by E. coli capable of colonizing the urinary tract, collectively known as uropathogenic E. coli (UPEC) (Zhang et al., 2002). From an evolutionary perspective, UPEC together with other extraintestinal pathogenic E. coli (ExPEC) belong to the E. coli phylogroups B2 and D, characterizing their specific adaptations to colonize and cause infections outside of the gut (Chen et al., 2013). Since the urinary tract presents a signficantly different environment than the gut, UPEC carry virulence factors very different from diarrheagenic E. coli (Kaper et al., 2004). For example, UPEC possess adhesins to attach to epithelial cells of the urinary tract to overcome the frequent flow of fluids (Oelschlaeger et al., 2002) and specific toxins for invading and replicating in the urinary tract (Mulvey, 2002). These known uropathogenic virulence factors presumably have multiple functions, as there is no direct correlation between these factors and UTI symptoms (Marrs et al., 2005). UPEC display a high diversity of genotypes and phenotypes (Zhang and Foxman, 2003, Landgren et al., 2005), suggesting that UPEC have multiple origins (Foxman and Brown, 2003, Wiles et al., 2008).
However, previous insights into the origins and spread of uropathogenecity were limited by their focuses on small regions of the bacterial genome that are well-conserved, such as genes used in mutlilocus sequence typing (MLST)(Marrs et al., 2005, Gibreel et al., 2012). These regions provide limited insight in the evolution of pathogenicity as they do not contain any of the virulence factors. Marrs et al. (2005) classified UPEC by grouping them into pathotypes based on virulence factors, analogous to the pathotypes for diarrheagenic E. coli (Nataro and Kaper, 1998). However, they did not find direct correlation between pathotype and clinical presentation. Other attempts of grouping UPEC by virulence factors also failed to identify a correlation between virulence factors and UTI symptoms (Tarchouna et al., 2013, Yun et al., 2014). These classification attempts suggest that UPEC virulence and genetic diversity cannot be captured by studying only a restricted set of genomic regions.
To allow a more complete understanding of the virulence and genetic diversity of bacterial strains, we examined full bacterial genomes in high resolution. To understand the evolution of uropathogenicity, we sequenced at over 190× coverage the genome of 19 E. coli strains isolated from UTI patients, 14 pathogenic strains from urine samples and 5 non-UTI-causing (“commensal” at the time of infection) rectal strains. We applied a de novo assembly-based algorithm to identify variants among the 19 strains, and constructed a whole-genome phylogeny based on these variants via a neighbor-joining algorithm.
In the whole-genome phylogeny, two commensal E. coli without typical combinations of pathogenicity genes formed the outgroup. This suggested that pathogenicity genes were present in infectious UPEC strains for a long time, with an estimated split from non-pathogenic E. coli over 32 million generations in the past. Even though our strains were collected in a small geographic area within a short period of time, we found high pairwise genomic diversity between any two strains of E. coli in our sample, which was incompatible with recent epidemics of a subset of strains.
To contrast the evolutionary signature of the strains with the evolution of individual uropathogenic virulence factors, we constructed gene trees of the three most common virulence factors in our sample. Comparing the whole-genome phylogeny to gene trees of uropathogenic factors, we observed that the virulence gene trees displayed a topology distinct from the whole-genome tree, suggesting that the whole-genome phylogeny could not capture the specific evolutionary history of virulence factors. We identified no excess horizontal gene transfer at these virulence factors, as indicated by the observation that the topology of the virulence gene trees were not more different from the whole-genome tree than the topology of random region gene trees were from the whole-genome tree. Hence the uropathogenicity in our strains was not the result of a recent adaptation. Instead, uropathogencity appeared to be a maintained ability in a subset of E. coli. These UPEC carried uropathogenicity genes for a long time, and they used such virulence opportunistically.
Methods
Study design
We selected 19 E. coli isolates from 14 subjects with UTI, including 14 UPEC isolates from urine samples and 5 non-UTI-causing (“commensal” at the time of infection) rectal E. coli. The 5 commensal strains were isolated from the same individuals and at the same time point as 5 of the UPEC isolates The samples were selected from a large collection of previously isolated strains from female patients attending the same clinic for UTI between 1996 and 2007. Sampling from the collection was based on pathotypes as defined by Marrs et al. (Marrs et al., 2005) based on common groupings of known uropathogenic factors (Table 1), in order to ensure a diversity of virulence factors in our sample. The 19 strains belonged to pathotypes 1 to 4 and pathotype 0, which is comprised of strains with no major groupings of uropathogenic factors. We employed a paired design, where each of the 5 commensal E. coli was isolated from an individual that also hosted one of the selected UPEC strains. We chose these pairs so that the commensal strain belonged to a different pathotype than the UPEC strain within each pair (Table 2).
Table 1.
Summary table of pathotype classification scheme, adapted from Marrs et al. (Marrs et al., 2005).
| Pathotype | Virulence factors |
|---|---|
| Pathotype 1 | cnf1, hly, prsGJ96 |
| Pathotype 2 | cnf1, hly, sfa |
| Pathotype 3 | aer, hly, papGAD/IA2 |
| Pathotype 4 | aer, kpsMT, ompT, drb |
| Pathotype 5 | kpsMT, ompT |
| Pathotype 0 | all remaining strains |
Pathotypes are assigned hierarchically to over 800 UPEC strains by examining pairwise association of 10 known virulence factors.
Table 2.
Samples description of the pilot study.
| Patient ID | Source | MLST Type | Pathotype | Code |
|---|---|---|---|---|
| 1 | Urine | ST 127 | 1 | 01U1 |
| 1 | Fecal | ST 12 | 2 | 01F2 |
| 2 | Urine | ST 12 | 1 | 02U1 |
| 2 | Fecal | ST 131 | 4 | 02F4 |
| 3 | Urine | ST 73 | 2 | 03U2 |
| 3 | Fecal | ST 73 | 1 | 03F1 |
| 4 | Urine | ST 144 | 3 | 04U3 |
| 4 | Fecal | ST 2731 | 0 | 04F0 |
| 5 | Urine | ST 404 | 4 | 05U4 |
| 5 | Fecal | ST 10 | 0 | 05F0 |
| 6 | Urine | ST 73 | 1 | 06U1 |
| 7 | Urine | ST 127 | 1 | 07U1 |
| 8 | Urine | ST 544 | 1 | 08U1 |
| 9 | Urine | ST 127 | 1 | 09U1 |
| 10 | Urine | ST 95 | 1 | 10U1 |
| 11 | Urine | ST 73 | 2 | 11U2 |
| 12 | Urine | ST 12 | 3 | 12U3 |
| 13 | Urine | ST 131 | 4 | 13U4 |
| 14 | Urine | ST 131 | 4 | 14U4 |
19 E. coli isolates were selected from 14 female patients attending the same clinic for UTI. 14 isolates were UPEC from urine sample and 5 were commensal E. coli from rectal swab sample, paired with one of the 14 UPEC from the same individual. We listed the MLST type and pathotype (Marrs et al 2005) of each strain. We labeled each strain by a four-digit code: first two digits represent individual host ID (01–14), the third digit represents UPEC from urine (U) sample or commensal E. coli from fecal (F) sample. The last digit represents the pathotype of the strain.
We regrew the 19 E. coli isolates from −80 °C stocks. We sequenced their genome using a single flow cell on Illumina HiSeq that produced 120 bp paired-end reads. The sequencing yield per sample ranged from 756 Mb to 1,328 Mb, totalling 19,535 Mb across all samples.
Variant calling
We employed a two-step, de novo assembly-based method (Cortex) to simultanteously reconstruct contigs and identify variants across multiple samples (Iqbal et al., 2012, Iqbal et al., 2013). This method is known to be conservative with high specificity (Young et al., 2012). Using a graph-building algorithm, Cortex constructs a colored de Bruijn graph from the sequence reads, where each color represents a sample. The resulting graph is error-cleaned by dynamically selecting a cleaning threshold from the coverage distribution. Divergence between samples exists as bubbles on the cleaned graph representation.
We employed the bubble calling algorithm in Cortex to detect variations between samples. We used a low k-mer (k=31) and a high k-mer (k=61) to build assembly graphs because low k-mers allow discovery of variants at relatively lower coverage, and large structural variations and genome complexity are more accessible at high k-mers (Iqbal et al., 2012). We combined variants called using the two k-mers into a joint call set. To get relative positions and to filter duplicate calls and overlapping sites, we aligned the assembled contigs, including each varying site and its flanking regions, with respect to each other. For the purpose of this study, we disregarded the complex variations including long segments of insertions, deletions or repeats and used only the single nucleotide polymorphisms (SNPs) for the following phylogenetic analyses.
We annotated all SNPs based on the genbank annotation of a uropathogenic E. coli refrence strain (UTI89), using the coordinate-only method in Cortex (Iqbal et al., 2013). We identified the phylogroup of each strain based on the presence and absence of three loci described in Clermont et al. (2000). In addition, we used this annotation to tabulate the presence and absence of 23 virulence factors (Spurbeck et al., 2012, Spurbeck and Mobley, 2013, Yun et al., 2014) in each strain.
Phylogenetic analyses
Using the SNPs identified from Cortex, we computed the pairwise sequence difference between samples and clustered them using a neighbor-joining algorithm (Saitou and Nei, 1987). We used Escherichia fergusonii (E. fergusonii) to root the phylogeny, since it is the closest species to E. coli (Touchon et al., 2009). To do so, we oriented the variants to E. fergusonii using the coordinate-only method in Cortex (Iqbal et al., 2013). In this way, variant discovery was independent of the choice of rooting or reference genome. To measure the confidence of the whole-genome phylogeny, we employed a bootstrap algorithm to resample the sequences of variants from the samples 10,000 times and obtain bootstrap values of the branches. We applied Phylip (Felsenstein, 2005) as neighbor-joining and bootstrapping algorithms. We studied clustering patterns on the phylogeny based on pathotype, host individual, and sequence type (ST) defined by the University College Cork E. coli scheme (Wirth et al., 2006). To test the signifiance of specific clustering patterns, we calculated the probability of a cluster given the tree topology, under the null hypothesis that the labeling of the tree is completely random; a small probability indicates that the cluster is unlikely to occur by chance.
To understand strain divergence times, we scaled the tree branches using a calibrated substitution rate of E. coli from Wielgoss et al. (2011). The rate was inferred directly from tracking the accumulation of synonymous substitutions via whole-genome sequencing of 19 E. coli strains in a 40,000-generation evolution experiment. We compared this calibration with alternative substitution rates presented in the earlier literature that were based on comparing sequences with known divergence times (Lecointre et al., 1998, Ochman et al., 1999). We categorized variants into synonymous and non-synonymous substitutions, and counted the number of synonymous and non-synonymous sites on the coding region, to estimate non-synonymous/synonymous rate ratio using a maximum likelihood method (Yang and Nielsen, 2000).
To contrast the evolution of the organism with the evolution of UTI virulence, we selected three UTI virulence factors: hly, aer, kpsMT (Marrs et al., 2005). Each was carried by over half of our sampled strains. We derived gene trees from the annotated variants called at each virulence factor and evaluated clustering by pathotype on these gene trees. The hly virulence factor consists of 4 genes: hlyA, hlyB, hlyC and hlyD, and the combined length is 7,281 bp. aer and kpsMT are 1,521 bp and 777 bp long, respectively. When constructing the respective gene tree, we considered only samples carrying the complete virulence factor. While kpsM is the definitive virulence factor for pathotype 4, we discarded one pathotype 4 strain (14U4) in this construction due to low sequencing coverage at the region. The first 200 bp of the 777-bp region were sequenced at less than 10-fold for this particular strain.
To compare a virulence factor’s gene tree with the whole-genome tree, we reconstructed the whole-genome phylogeny based only on the samples carrying the virulence factor. We scaled branch lengths by the total number of variants on a tree. We then measured the similarity of the gene tree and the whole-genome tree using a topological score, generated by a branch-matching algorithm that searches for the optimal one-to-one transformation between two trees (Nye et al., 2005). We contrasted the similarity score of each gene tree with an empirical distribution of similarity scores of trees containing the same number of leaves and same number of variants as the virulence gene tree. We generated this empirical distribution by randomly drawing sets of the same number of consecutive variants as each gene tree and generating trees based on these sets of varaints. We then calculated the topological similarity score of each random tree to the whole-genome tree, which gave us an empirical distribution of similarity scores. A score at the extremes of the empirical distribution indicates that the gene tree is significantly more different from or more similar to the whole-genome tree than random regions of the genome.
Results
Whole-genome phylogeny
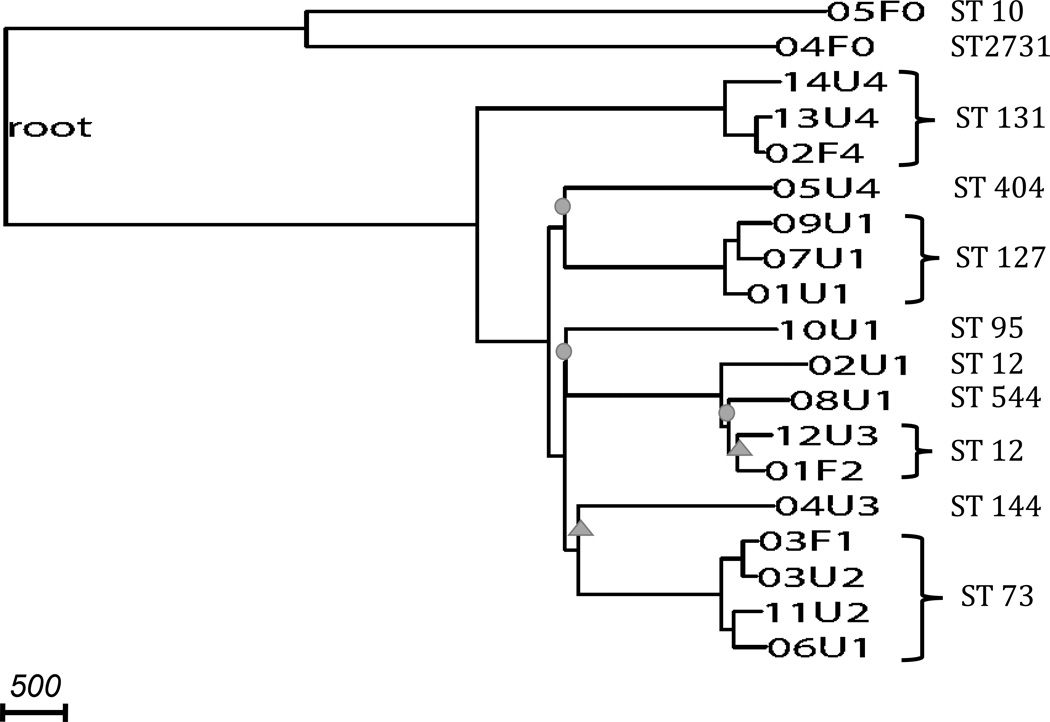
Using de novo assembly-based variant calling methods, we identified 68,396 SNPs with a transition-to-transversion ratio of 2.73. All our 19 strains belonged to the phylogroup B2. We oriented 24,568 of the variant set to the E. fergusonii outgroup sequence coordinates and constructed a rooted phylogeny (Figure 1). Most splits on this whole-genome phylogeny had bootstrap values of 100%, while two splits had 95–100% bootstrap values, and three had 65–95% bootstrap values.
Figure 1. Phylogeny constructed from whole-genome assembly-based variants.
The phylogeny is rooted by E. fergusonii (long branch not shown). Nodes are labeled by the sample codes as listed in Table 2. All strains are of phylogroup B2. Branches with circles represent bootstrap values, and branches with triangles represent bootstrap values. All unmarked branches have bootstrap value. Scale shows length on branches representing 500 pairwise sequence differences.
Applying the E. fergusonii gene annotation to our variant set, we identified 11,216 synonymous mutations (45.7% of the variants), and counted 963,414 synonymous sites on the oriented genome. The maximum likelihood estimate of the ratio of the number of non-synonymous substitutions per non-synonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks) was 0.54, indicating purifying selection consistent with previous findings (Jordan et al., 2002). Using an estimated substitution rate of 8.9×10−11 per base pair per generation, based on the laboratory evolution of E. coli (Wielgoss et al., 2011), the evolutionary time elapsed on the entire phylogeny was over 130 million generations. Based on this calibration, we expected 1 synonymous mutation per 11,600 generations. Alternative substitution rate estimates based on sequences with known divergence times were 5.2×10−11 per base pair per generation in Lecointre et al. (1998), and 3×10−11 per base pair per generation in Ochman et al. (1999), which led to 1.5- to 3-fold shorter evolutionary time.
Two non-UTI-causing (commensal) strains, both belonging to pathotype 0, formed the outgroup on the rooted phylogeny (Figure 1) (p=0.0058). The split between this outgroup and the remaining phylogeny represents the time of divergence of UPEC strains from commensal strains ocurred; we estimated a split time of 32 million generations ago.
We observed that the clustering of strains on our whole-genome phylogeny did not correspond to pathotype classification. Strains of pathotypes 1, 2 and 3 showed no distinct subclades and together formed a single large cluster, regardless of whether the strain was a commensal or uropathogenic E. coli (Figure 1). Similarly, applying the grouping methods of UPEC based on presence of several virulence genes described in Tarchouna et al. (2013) (Grouping 1) and Yun et al. (2014) (Grouping 2), we observed that none of the groups fell completely and distinctively into subclades (Figure 1, Supplementary Figure 1). In pathotype and Grouping 2 classifications, each had one type with four strains where three strains formed a significant subclade (p=0.0041), but the remaining strain of the same type was far away from the subclade on the phylogeny. In the pathotype classification, three pathotype 4 strains of ST 131 clustered, but the remaining pathotype 4 strain (05U4) in our sample had a pairwise sequence difference of over 4,700 with the other pathotype 4 strains. The split of 05U4 with the other pathotype 4 subclade happened over 10 million generations ago. Among the three strains that clustered, the shortest external branch leading to 02F4 still represented over 320,000 generations, indicating long divergence times between pathotype 4 strains (Figure 1). Similarly, based on Grouping 2 classification, three type 6 strains of ST 127 clustered, with a mean pairwise sequence difference of over 500. However, the remaining group 6 strain (02U1) had a pairwise sequence difference of over 3,800 with the other group 6 strains (Supplementary Figure 1). The split of 02U1 with the other type 6 subclade happened over 7 million generations ago. Moreover, when tabulating the virulence genes present in each strain, we found that strains from the same ST did not necessarily carry the same set of virulence genes (Supplementary Table 1).
To investigate if multilocus sequence typing (MLST) based on seven housekeeping E. coli loci was consistent with our whole-genome phylogeny, we identified the ST of each strain in our sample (Table 3). While most strains in our sample were singletons in the ST classification, four STs had three or more strains in our sample, namely, ST 12, ST 73, ST 127 and ST 131 (Table 3). We observed that organinisms with the same ST mostly formed consistent subclades on the phylogeny (Figure 1), with the exception of the ST 12 cluster, which also contained one ST 544 strain. However, all splits defining this cluster had less than 100% bootstrap value. Nonetheless, strains within each ST still had remarkable diversity (Table 3): For ST 73, ST 127 and ST 131, each pair of strains within its respective ST had on average over 500 differences, reflecting a divergence of >360,000 generations. ST 127 strains displayed the highest overall similarity with an average of 501.3 pairwise differences. The second most similar type was ST 73 with an average of 554.5 pairwise differences. ST 131 strains had an average of 597.3 pairwise differences. ST 12 strains were more diverse, with an average of 907 pairwise differences.
Table 3.
Pairwise sequence differences of strains belonging to the same ST
| ST 131 | ST 127 | ST 73 | ST 12 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14U4 | 13U4 | 02F4 | 09U1 | 07U1 | 01U1 | 03F1 | 03U2 | 11U2 | 06U1 | 02U1 | 12U3 | 01F2 | ||
| ST 131 | 14U4 | 0 | 829 | 785 | ||||||||||
| 13U4 | 0 | 178 | ||||||||||||
| 02F4 | 0 | |||||||||||||
| ST 127 | 09U1 | 0 | 462 | 497 | ||||||||||
| 07U1 | 0 | 545 | ||||||||||||
| 01U1 | 0 | |||||||||||||
| ST 73 | 03F1 | 0 | 237 | 638 | 712 | |||||||||
| 03U2 | 0 | 563 | 681 | |||||||||||
| 11U2 | 0 | 496 | ||||||||||||
| 06U1 | 0 | |||||||||||||
| ST 12 | 02U1 | 0 | 119 | 1015 | ||||||||||
| 12U3 | 0 | 512 | ||||||||||||
| 01F2 | 0 | |||||||||||||
Finally, when evaluating the matched pairs of one commensal and one UPEC strain sampled from the same individual, we observed that only one pair clustered with a pairwise difference of 237 variants. The other four pairs were located very far apart on the tree (Figure 1); they did not cluster in the same subclade, indicating no significant excess of clustering (p=0.19).
Phylogeny of virulence factors
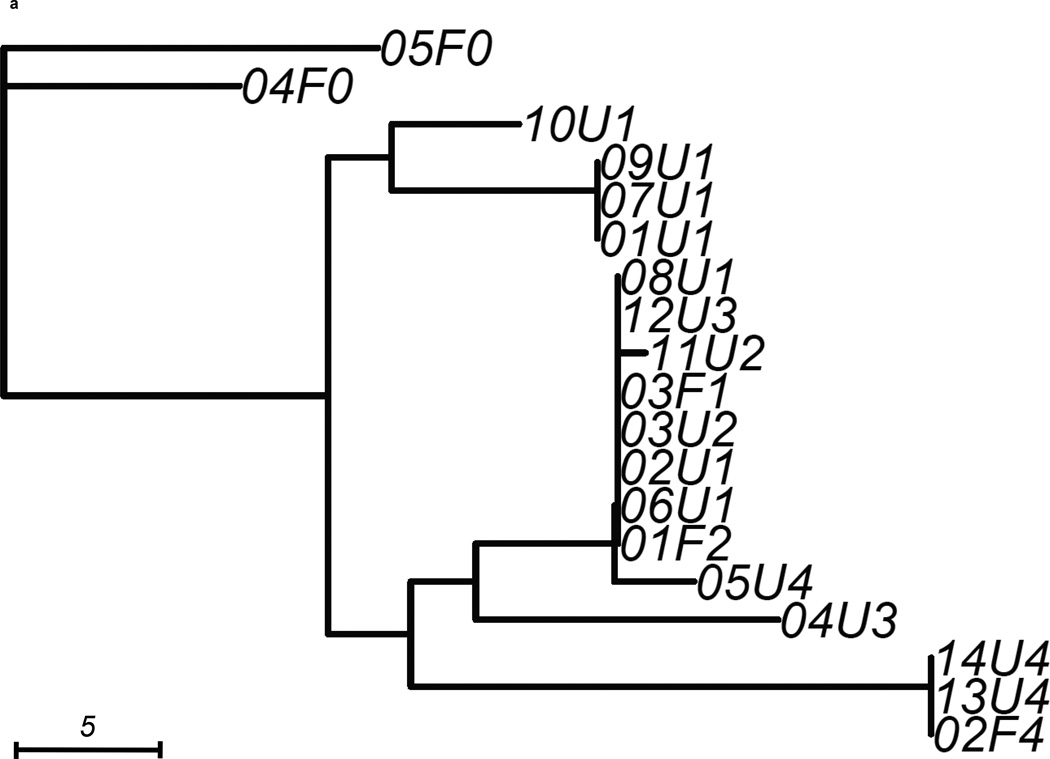
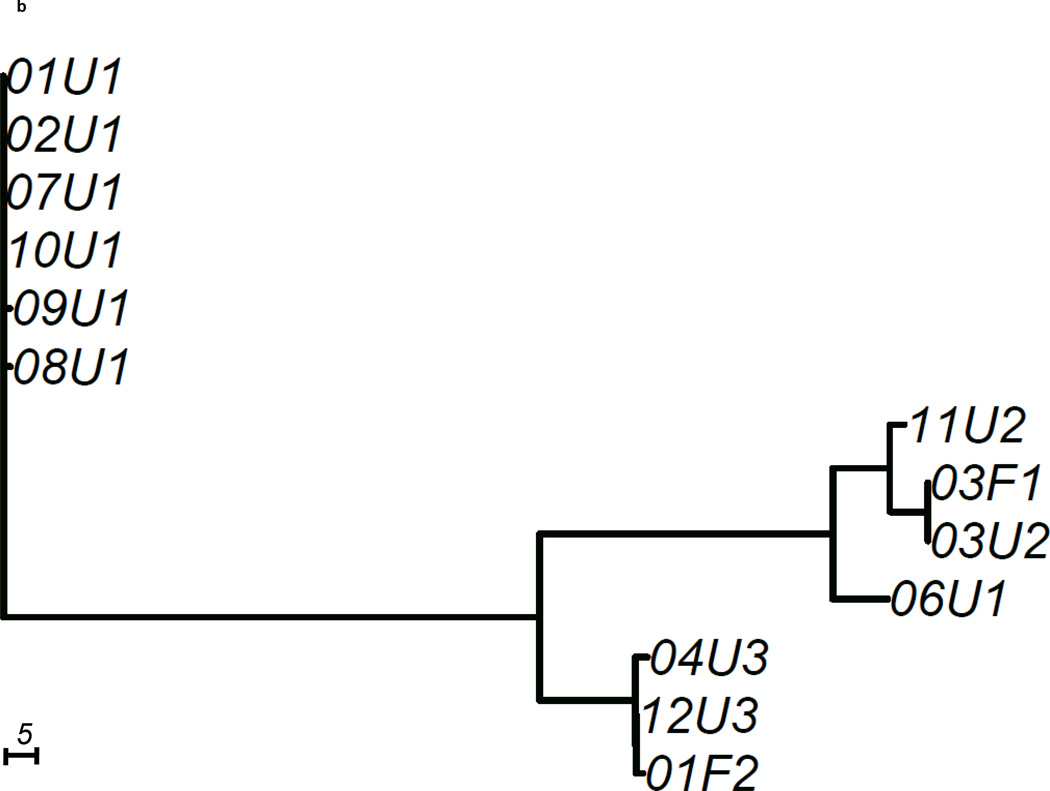
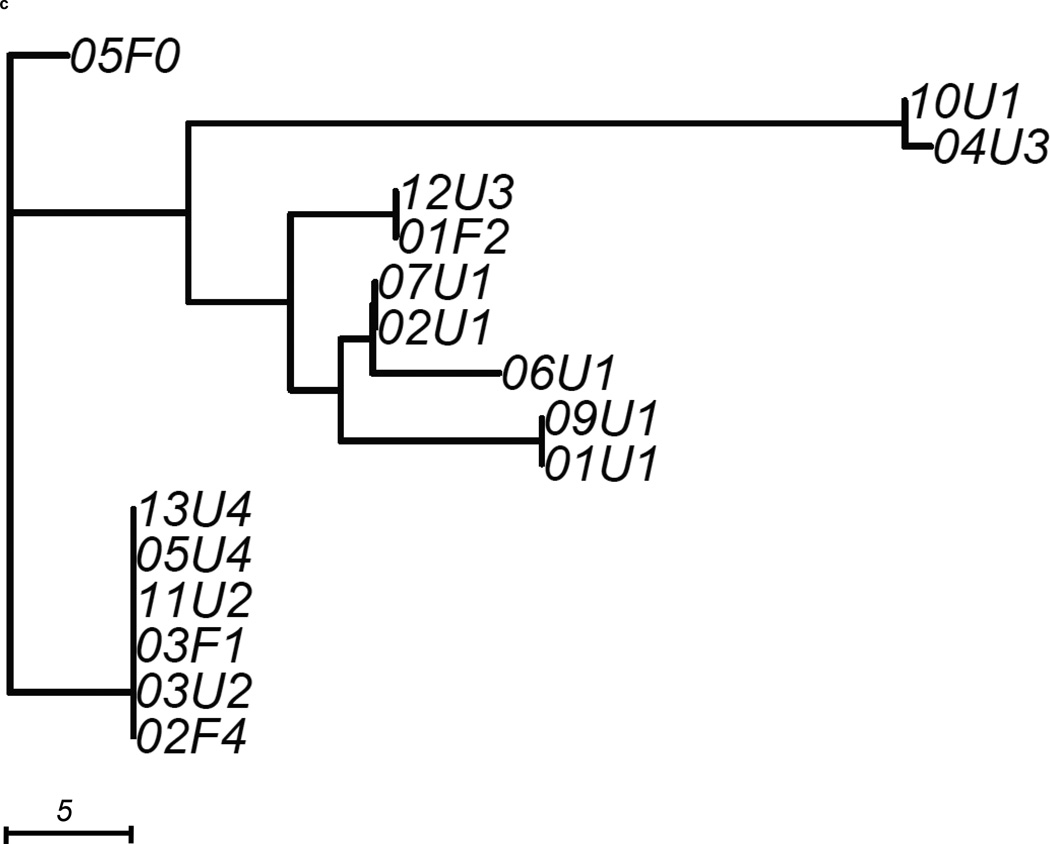
We constructed 3 gene trees from known uropathogenic virulence factors (Marrs et al., 2005) that are present in multiple strains in our sample: aer, hly and kpsMT (Figure 2). Each gene tree consisted of a different number of tree leaves, because not all virulence factors were found in all strains: All 19 strains carried aer, 13 strains carried the complete segment of hly, while 16 strains carried kpsMT (Table 4). After scaling branch lengths by the number of mutations on a tree, we compared each gene tree to a subtree of the whole-genome phylogeny consisting of the corresponding subset of strains. For this comparison, we applied a topological score (Nye et al., 2005) which summarizes the percentage of topological similarity between branches of two trees.
Figure 2. Unrooted trees derived from three selected virulence factors: (a) aer, (b) hly, and (c) kpsMT for uropathogenic and commensal E. coli.
Each gene tree consists of a different number of tree leaves, as not all virulence factors occurred on all strains. Scale shows length on branches representing 5 pairwise sequence differences.
Table 4.
Virulence gene trees: gene length, number of carriers (out of 19 samples), topological similarity scores compared to whole-genome tree and p-values of these scores generated from the empirical distribution of scores from random trees.
| Virulence gene | Gene length | Number of carriers | Topological Similarity Score | p-value |
|---|---|---|---|---|
| aer | 1,521 bp | 19 | 0.635 | 0.318 |
| hly | 7,281 bp | 13 | 0.565 | 0.185 |
| kpsMT | 777 bp | 16 | 0.516 | 0.209 |
The aer gene tree, containing all 19 strains, possessed similar structural features as the whole-genome tree (Figure 2a) with a topological score of 0.635 (Table 4). The difference between the two trees was best illustrated by the two strains 04U3, 05U4 that segregated differently in this gene tree than on the whole-genome phylogeny. Some strains carried identical copies of the virulence gene. For example, pathtoype 1 UPEC strains (01U1, 07U1, 09U1) displayed no pairwise sequence difference at this gene. The two fecal strains with virulence factors (03F1, 01F2) were also identical at this gene, so were three other UPEC (02U1, 03U2, 06U1). When we assessed similarity by comparing the topology score against to the empirical distribution of scores, we saw no signal that the similarity between the aer gene tree and whole-genome tree was higher or lower than other trees containing the same number of variants (p=0.318, Table 4).
The hly gene tree showed more differences from the whole-genome tree with a topological score of 0.565 (Table 4). Only the 13 strains of pathotypes 1,2 and 3 carried the complete segment of hly (Figure 2b). On the hly gene tree (Figure 2b), we observed that four pathotype 1 UPEC strains (01U1, 02U1, 07U1, 10U1) carried the identical copy of hly as the UTI89 reference strain. Two other pathotype 1 strains (08U1, 09U1) had a hly copy with only one base different from the UTI89 hly. The hly regions of the remaining two pathotype 1 strains were significantly different from the other pathotype 1 hly regions. 03F1 and 06U1 each displayed over 100 pairwise sequence differences at hly when compared to the UTI89 hly. These two hly were more similar to the pathotype 2 hly, as a result formed a cluster on the hly tree. The hly region of the pathotype 2 fecal strain were very similar to those of pathotype 3 strains, with 1 and 3 pairwise differences respectively. While the similarity score of the hly gene tree to whole-genome tree (0.565) was lower than that of the aer gene tree (0.635), there was no strong signal that the hly tree was less similar to the whole-genome tree than random genomic regions (p=0.185, Table 4).
The kpsMT gene tree consisted of 16 strains and was the least similar to the whole-genome phylogeny among the three virulence factors studied, with a topological score of 0.516 (Figure 2c, Table 4). This factor was completely absent in a commensal pathotype 0 strain (04F0) and one pathotype 1 UPEC strain (08U1). The resulting gene tree showed that four UPEC and two commensal strains were identical at this gene The remaining strains displayed considerable diversity at this gene, as indicated by the longer branches. The longest branch leading to the 10U1 and 04U3 subclade contained 28 mutations. Notably, strains that clustered closely together in the previous two gene trees appeared to be further apart on this gene tree. However, the kpsMT gene tree was not significantly less similar to the whole-genome tree than random genomic regions (p=0.209, Table 4).
Discussion
We studied the evolution of pathogenicity in uropathogenic E. coli (UPEC) with the goal of understanding the origin and spread of UPEC in the context of urinary tract infections. We sampled strains from a large collection of E. coli isolates with well-characterized virulence factors (Marrs et al., 2005), in order to expand the diversity of virulence factors in our sample. This approach is different from most existing studies where sampling is based on clinical visits (Tarchouna et al., 2013, Yun et al., 2014). This study design allowed us to study a broader spectrum of virulence factors in order to understand the evolution of uropathogenicity.
Using whole-genome deep-sequencing, we explored whole bacterial genomes at high resolution, allowing more detailed analyses than pathotype or MLST schemes that study only small regions of the genome. We employed a multi-sample de novo assembly algorithm Cortex that simutaneously assembles genomes and calls variants (Iqbal et al., 2012). This method calls variants independently of a reference genome. Hence, the variant calls are unaffected by the choice of reference sequence, making this approach well-suited to a sample with high diversity such as ours. The variant calls generated from Cortex are known to be conservative with high specificity (Young et al., 2012). These high quality whole-genome variants allowed a more accurate investigation of the evolutionary pathways of uropathogenicity and the degree of diversity among strains.
Our phylogenetic analyses constrasting UPEC with non-pathogenic E. coli showed that their divergence happened over 32 million generations ago, which is equivalent to 107,000 to 320,000 years, assuming the E. coli had 100–300 generation per year (Ochman et al., 1999). Alternative estimates of substitution rates gave qualitatively consistent results, with an estimated 10.6–18.7 million generations on the whole-genome phylogeny. Between pathogenic strains, the whole-genome pariwise diversity was high, corresponding to a long divergence history of over 130 million generations, or 0.4–1.3 million years. Both of these estimates from the whole-genome phylogeny showed that within the small geographic region of our sample collection, UTI were caused by strains of multiple origins. In addition, commensal and UPEC strains from the same individual were as different from each other as from other strains in the sample, suggesting that the infection was unlikely caused by gut E. coli strains that recently acquired uropathogenicity factor within the human host.
Our phylogenetic analysis of the entire E. coli genome allowed us to evaluate more basic methods of E. coli classification such as the MLST scheme and pathotype groupings based on virulence factors. The pathotype classification we used for sampling did not capture the overall relationship of strains. In particular, pathotypes 1, 2 and 3 did not form distinctive clusters on the whole-genome phylogeny. When we applied to our sample alternative groupings of UPEC strains based on other distinct sets of virulence factors (Tarchouna et al., 2013, Yun et al., 2014), we also observed that most groups did not cluster well on the whole-genome phylogeny. For the pathotype and group that had three out of four strains forming a significant subclade, we observed high diversity among the three strains that clustered, and a deep split between the subclade and the remaining strain of the same type. Therefore, classification based on presence and absence of virulence factors did not appear to be meaningful for understanding the evolutionary history of UPEC strains.
On the other hand, classification based on the traditional MLST scheme did generally capture the evolutionary relationship of strains. However, our whole-genome phylogeny identified high diversity among strains that were classified into the same sequence type, something that was previously not well-studied. The divergence time of particular clusters, for example the pathotype 4 ST 131 cluster, was longer than suggested by previous studies (Clark et al., 2012, Nicolas-Chanoine et al., 2014). Long branch lengths of the ST 131 cluster reflected ancient origins and high diversity within the ST. Consistent with our findings, a recent study using a phenotypic microarray showed that ST 131 was not a distinct lineage of ExPEC (Alqasim et al., 2014). We observed a similar level of diversity in other sequence types, with substantial variation among strains of the same ST of over 100 pairwise differences. Long divergence times among strains of the same ST suggests that they are not clonal, as they evolve to accumulate large number of genomic differences over time. Moreover, by tabulating the presence and absence of 23 potential uropathogenic virulence factors in our strains, we observed that strains of the same ST carried different sets of virulence factors. Therefore, classifying UPEC by sequence type is not sufficient for drawing inferences on the presence of UPEC virulence factors.
We further explored the evolutionary pathways of individual uropathogenic virulence factors by constructing gene trees of three virulence factors that were the most common in our sample. We aimed to identify evidence for horizontal gene transfer as horizontal gene transfer is the major mechanism for non-pathogenic E. coli to aquire uropathogenicity. If uropathogenic E. coli strains acquired pathogenicity via elevated rates of horizontal gene transfer, or preferential selection of horizontally transferred virulence genes, the corresponding gene tree would display a significantly different topology from the whole genome tree. Therefore, we constrasted the topolgy of the gene trees and the whole-genome phylogeny to see if virulence genes displayed distinct evolutionary pathways. We found that each virulence gene tree had an evolutionary pathway distinct from the whole-genome phylogeny, indicated by the 50–65% topological similarity scores. Based on the empirical distribution of similarity scores of random gene regions, the virulence gene trees did not have extreme similarity scores, meaning these uropathogenic genes were not significantly more similar to the whole-genome phylogeny than other genomic regions. This suggests no signal of excess horizontal gene transfer and no selective advantage at the uropathogenic genes than elsewhere in the genome.
In summary, by quantifying the diversity of UPEC strains using whole-genome deep-sequencing and constrasting with commensal E. coli, we showed that UPEC had a long evolutionary history since their divergence from non-UTI-causing commensal E. coli. Our study illuminated the development of UTI and showed that UPEC are opportunistic, conserving uropathogenic virulence factors without signals of preferential selection or increased rates of horizontal gene transfer. Our results indicated that the phylogenetic relationship of UPEC provided only limited information about the presence of virulence factors and thus suggested that closely related UPEC may have dissimilar uropathogenic phenotypes. Further extensive sequencing of UPEC and commensal E. coli will allow deeper understanding of the genetic signals and mechanisms driving the epidemiology of uropathogenicity.
Supplementary Material
Highlights.
-
-
We sequenced 14 uropathogenic E. coli and 5 commensals at >190×.
-
-
We found deep split between uropathogenic and commensal E. coli (>32 million generations).
-
-
High between-strain diversity showed no signal of epidemics in the sampled area.
-
-
High diversity in each ST and pathotype suggested multiple origins of pathogenicity.
-
-
We detected no selective advantage of virulence factors over other genomic regions.
Acknowledgements
We thank Carl Marrs, Daniel Falusch, Hyun Min Kang and Kirsten Herold for helpful input related to this work. This research was supported by funds from the Center for Molecular and Clinical Epidemiology of Infectious Diseases at the University of Michigan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author's contributions
YL carried out the analysis and wrote the manuscript. LZ, SZ and BF designed the study and critically revised the manuscript. BF provided the samples. All authors read and approved the final manuscript.
References
- Alqasim A, Emes R, Clark G, Newcombe J, La Ragione R, McNally A. Phenotypic microarrays suggest Escherichia coli ST131 is not a metabolically distinct lineage of extra-intestinal pathogenic E. coli . PLoS One. 2014;9:e88374. doi: 10.1371/journal.pone.0088374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Wu M, Henderson JP, Hooton TM, Hibbing ME, Hultgren SJ, Gordon JI. Genomic diversity and fitness of E. coli strains recovered from the intestinal and urinary tracts of women with recurrent urinary tract infection. Sci. Transl. Med. 2013;5:184ra60. doi: 10.1126/scitranslmed.3005497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark G, Paszkiewicz K, Hale J, Weston V, Constantinidou C, Penn C, Achtman M, McNally A. Genomic analysis uncovers a phenotypically diverse but genetically homogeneous Escherichia coli ST131 clone circulating in unrelated urinary tract infections. J. Antimicrob. Chemother. 2012;67:868–877. doi: 10.1093/jac/dkr585. [DOI] [PubMed] [Google Scholar]
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000;66:4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.69. 2005 [Google Scholar]
- Foxman B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 2002;113:5–13. doi: 10.1016/s0002-9343(02)01054-9. [DOI] [PubMed] [Google Scholar]
- Foxman B, Brown P. Epidemiology of urinary tract infections: transmission and risk factors, incidence, and costs. Infect. Dis. Clin. North Am. 2003;17:227–241. doi: 10.1016/s0891-5520(03)00005-9. [DOI] [PubMed] [Google Scholar]
- Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, Upton M. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. Br. Soc. Antimicrob. Chemo. 2012;67:346–356. doi: 10.1093/jac/dkr451. [DOI] [PubMed] [Google Scholar]
- Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G. de novo assembly and genotyping of variants using colored de Bruijn graphs. Nat. Genet. 2012;44:226–232. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal Z, Turner I, McVean G. High-throughput microbial population genomics using the Cortex variation assembler. Bioinformatics. 2013;29:275–276. doi: 10.1093/bioinformatics/bts673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Microevolutionary genomics of bacteria. Theor. Popul. Biol. 2002;61:435–447. doi: 10.1006/tpbi.2002.1588. [DOI] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli Nature Rev. Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Landgren M, Odén H, Kühn I, Österlund A, Kahlmeter G. Diversity among 2481 Escherichia coli from women with community-acquired lower urinary tract infections in 17 countries. J. Antimicrob. Chemother. 2005;55:928–937. doi: 10.1093/jac/dki122. [DOI] [PubMed] [Google Scholar]
- Lecointre G, Rachdi L, Darlu P, Denamur E. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol. Biol. Evol. 1998;15:1685–1695. doi: 10.1093/oxfordjournals.molbev.a025895. [DOI] [PubMed] [Google Scholar]
- Marrs CF, Zhang L, Foxman B. Escherichia coli mediated urinary tract infections: are there distinct uropathogenic E. coli (UPEC) pathotypes? FEMS Microbiol. Lett. 2005;252:183–190. doi: 10.1016/j.femsle.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Mulvey MA. Adhesion and entry of uropathogenic Escherichia coli Cell. Microbiol. 2002;4:257–271. doi: 10.1046/j.1462-5822.2002.00193.x. [DOI] [PubMed] [Google Scholar]
- Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 2014;27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye TMW, Liò P, Gilks WR. A novel algorithm and web-based tool for comparing two alternative phylogenetic trees. Bioinformatics. 2005;22:117–119. doi: 10.1093/bioinformatics/bti720. [DOI] [PubMed] [Google Scholar]
- Ochman H, Elwyn S, Moran NA. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. U. S. A. 1999;96:12638–12643. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlaeger TA, Dobrindt U, Hacker J. Virulence factors of uropathogens. Curr. Opin. Urol. 2002;12:33–38. doi: 10.1097/00042307-200201000-00007. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Spurbeck RR, Mobley HL. Uropathogenic Escherichia coli. Escherichia coli: Pathotypes and Principles of Pathogenesis. 2013;275 [Google Scholar]
- Spurbeck RR, Dinh PC, Jr, Walk ST, Stapleton AE, Hooton TM, Nolan LK, Kim KS, Johnson JR, Mobley HL. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 2012;80:4115–4122. doi: 10.1128/IAI.00752-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchouna M, Ferjani A, Ben-Selma W, Boukadida J. Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. International Journal of Infectious Diseases. 2013;17:e450–e453. doi: 10.1016/j.ijid.2013.01.025. [DOI] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguénec C, Lescat M, Mangenot S, Martinez-Jéhanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Médigue C, Rocha EPC, Denamur E. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS genetics. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielgoss S, Barrick JE, Tenaillon O, Cruveiller S, Chane-Woon-Ming B, Medigue C, Lenski RE, Schneider D. Mutation Rate Inferred From Synonymous Substitutions in a Long-Term Evolution Experiment With Escherichia coli. G3 (Bethesda) 2011;1:183–186. doi: 10.1534/g3.111.000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles TJ, Kulesus RR, Mulvey MA. Origins and virulence mechanisms of uropathogenic Escherichia coli . Exp. Mol. Pathol. 2008;85:11–19. doi: 10.1016/j.yexmp.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Young BC, Golubchik T, Batty EM, Fung R, Larner-Svensson H, Votintseva AA, Miller RR, Godwin H, Knox K, Everitt RG, Iqbal Z, Rimmer AJ, Cule M, Ip CL, Didelot X, Harding RM, Donnelly P, Peto TE, Crook DW, Bowden R, Wilson DJ. Evolutionary dynamics of-Staphylococcus aureus during progression from carriage to disease. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4550–4555. doi: 10.1073/pnas.1113219109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun KW, Kim HY, Park HK, Kim W, Lim IS. Virulence factors of uropathogenic Escherichia coli of urinary tract infections and asymptomatic bacteriuria in children. Journal of Microbiology, Immunology and Infection. 2014;47:455–461. doi: 10.1016/j.jmii.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Zhang L, Foxman B. Molecular epidemiology of Eschichia coli mediated urinary tract infections. Front. Biosci. 2003;8:e235–e244. doi: 10.2741/1007. [DOI] [PubMed] [Google Scholar]
- Zhang L, Foxman B, Marrs C. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J. Clin. Microbiol. 2002;40:3951–3955. doi: 10.1128/JCM.40.11.3951-3955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.