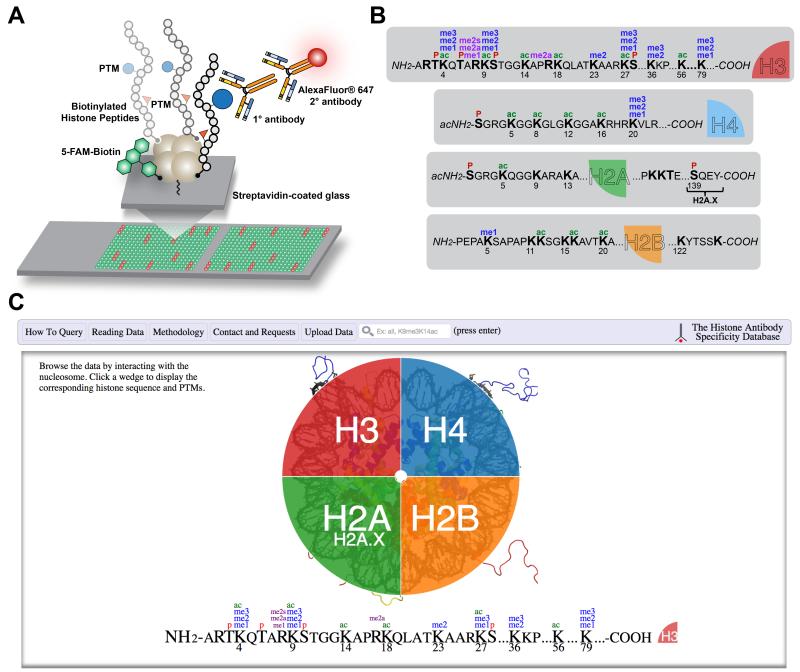
Figure 1. Histone antibody characterization by peptide microarray and web interface of the Histone Antibody Specificity Database.
(A) Cartoon depicting the stepwise procedure for peptide microarraying of an antibody from peptide immobilization to antibody hybridization and visualization. Positive interactions are visualized as red fluorescence. All printed spots show green fluorescence from the biotinylated fluorescein (5-FAM) printing control. (B) Posttranslational modifications and their combinations on human histones for which antibodies screened to publication date (Table S2) are marketed to recognize. Lysine acetylation (ac); Lysine methylation (me1/2/3*); Arginine methylation (me1/2a/2s**); Serine and threonine phosphorylation (P). *Histone lysine methylation occurs in three forms (mono-, di-, and trimethylation). **Arginine methylation also occurs in three forms (monomethylation, symmetric, and asymmetric dimethylation). (C) Screenshot of the home page for www.histoneantibodies.com, an interactive java-based web resource displaying histone antibody specificity data from peptide microarray experiments. Users can query data by entering strings in the search bar and by clicking on their histone and posttranslational modification of interest.