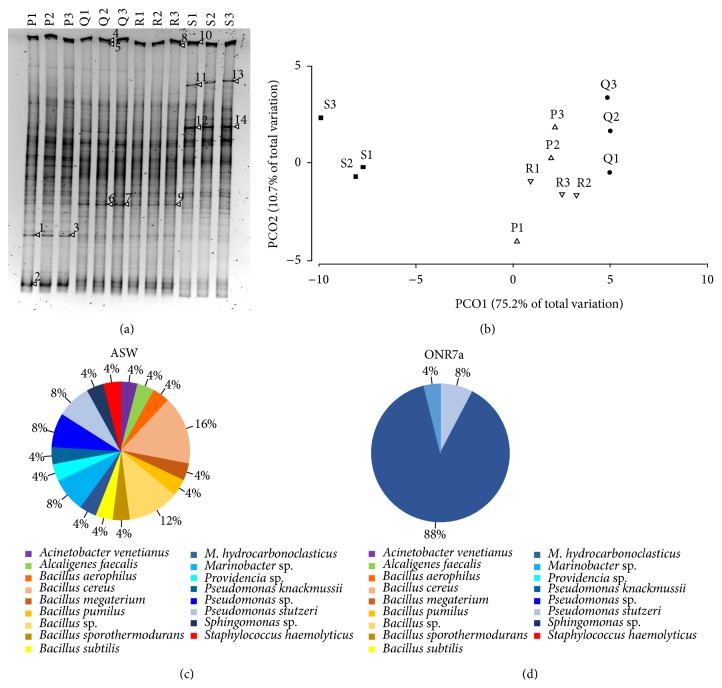
Figure 4.
Cultivation dependent and independent analyses of the bacterial communities at El-Max district. (a) DGGE analysis performed of the 16S rRNA on the sediment metagenome. The numbers in the name of the samples represented the three analyzed replicates. Bands numbered have been excised from the gel and their DNA content has been sequenced (results are reported in Table 5). (b) Principal component analysis based on the DGGE profiles of the 16S rRNA gene in the sediments. (c) Identification and relative abundance of the bacteria isolated on ONR7a medium. (d) Identification and relative abundance of the bacteria isolated on ASW medium.