Abstract
Pseudomonas syringae is a phytopathogenic bacterium widely spread on terrestrial plants. Sulfodiaminophosphinyl tripeptide Phaseolotoxins (PHTs), produced by P. syringae pv. phaseolicola and P. syringae pv. actinidiae, represent a kind of antimetabolic phytotoxins. PHTs inhibit host cell Ornithine transcarbamylase (OTCase) activity and induce Arginine auxotrophic phenotype. The biosynthesis of PHT is temperature dependent, being optically produced at around 18 °C, while blocked above 28 °C. PHT resistant OTCase ArgK acts as a functional replacement of housekeeping OTCase ArgF, which is the acting target of PHT, to confer PHT producers with self-resistance. It was postulated that argK might be regulated directly by a PHT biosynthetic precursor and indirectly by temperature with an unknown manner. Neither transcriptional regulator nor thermal regulation related protein encoding gene was detected from PHT biosynthetic gene cluster. The tripeptide, Cit-Ala-hArg, was identified to be a by-product of PHT biosynthetic pathway in this report. Formation of Cit-Ala-hArg was catalyzed by ArgK with tripeptide Orn-Ala-hArg and carbamyl phosphate as substrates. It showed that ArgK not only provided alternative Arginine source as reported previously, but also controlled the production of PHTs by converting PHT biosynthetic precursors to nontoxic Cit-Ala-hArg reservoir for producers’ self-defense.
Pseudomonas syringae (P. syringae) is a group of phytopathogenic bacterial species which include more than 50 pathovars. P. syringae spreads on most of the terrestrial plants and induces a wide variety of diseases1,2. Phosphorotriamidate natural product Phaseolotoxins (PHTs) producers, P. syringae pv. phaseolicola and P. syringae pv. actinidiae, cause halo blight disease on beans and bacterial canker on kiwifruits respectively3,4,5. PHTs [Nδ(N’-sulfo-diaminophosphinyl)-ornithyl-alanyl-homoarginine] were readily degraded by nonspecific peptidases in planta to produce compound Nδ (N’-sulfo-diaminophosphinyl)-ornithine (PSOrn)6,7,8. PSOrns are irreversible inhibitor of ornithine transcarbamylase (OTCase) attributed to the chemical structure similarity with tetrahedral intermediates in OTCase mechanism6. OTCase catalyzes the formation of Citrulline with Ornithine and carbamyl phosphate as substrates. Inhibition of OTCase blocks the biosynthesis of Arginine in vivo and leads to a reduction of protein synthesis9. In addition, PHTs are competitive inhibitors to mammalian and bacterial OTCase, including the OTCase ArgF of PHT producing strain P. syringae pv. phaseolicola 1448A10,11,12. So, PHT producing Pseudomonas cells must employ certain approaches to protect themselves from being killed by their own second metabolic products.
PHT resistant OTCase ArgK, encoded by argK, is a well-known self-resistance conferring element to PHT producers13,14,15. In P. syringae pv. phaseolicola cells, ArgK acts as a functional replacement of housekeeping OTCase ArgF to provide an alternative Arginine source whenever ArgF is inhibited by PHTs3,16. PHT biosynthetic gene cluster contains 23 genes (24.8 kb), which organized into five transcriptional units, two monocistronic units (argK and phtL) and three operons17,18,19. Biosynthesis of PHT is temperature dependent, being optimized between 18 °C and 20 °C, while blocked above 28 °C20,21. Resistant gene argK expressed at both 18 °C and 28 °C. However its transcriptional level was much lower at 28 °C17. It was postulated that argK might be regulated directly by a PHT biosynthetic precursor and indirectly by temperature. A protein-binding DNA motif has been found out in argK promoter region, while no repressor protein involved in thermoregulation of PHT biosynthesis has been identified so far22,23,24.
Gene phtL expressed at both 18 °C and 28 °C, and showed no quantity difference at transcriptional level17. PhtL, the product of phtL, is a bidomain enzyme which shows distantly similarity to pyruvate phosphate dikinase (PPDK) and phosphoenolpyruvate synthase (PS). It has been proposed that phtL played a regulatory function in PHT biosynthesis17,25. The amidinotransferase homolog encoded by amtA, was proposed to catalyze the conversion of Lysine to homoarginine (hArg) using Arginine as guanidyl group donors26. Two ATP grasp family peptide ligase encoding genes, phtQ and phtU were recognized and deduced to catalyze the formation of amido bonds of PHT peptide scaffolds Orn-Ala-hArg27. N-Sulfodiaminophosphinyl groups attach to the delta amino group of Ornithine residues give the chemical structure of PHTs. The chronological order of individual steps involved in PHT biosynthesis and the three Nitrogen-Phosphorus bonds (N-P bonds) biosynthetic mechanism remain poorly understood.
Here, precursor ion scan (PIS) mass spectrometry, feeding experiments and enzyme assay were employed in the identification of PHT biosynthetic pathway by-products, tripeptides Cit-Ala-hArg, which indicating a dual role of ArgK for PHT producers self-defense.
Results
PIS mass spectrometry to screen for PHT biosynthetic intermediates
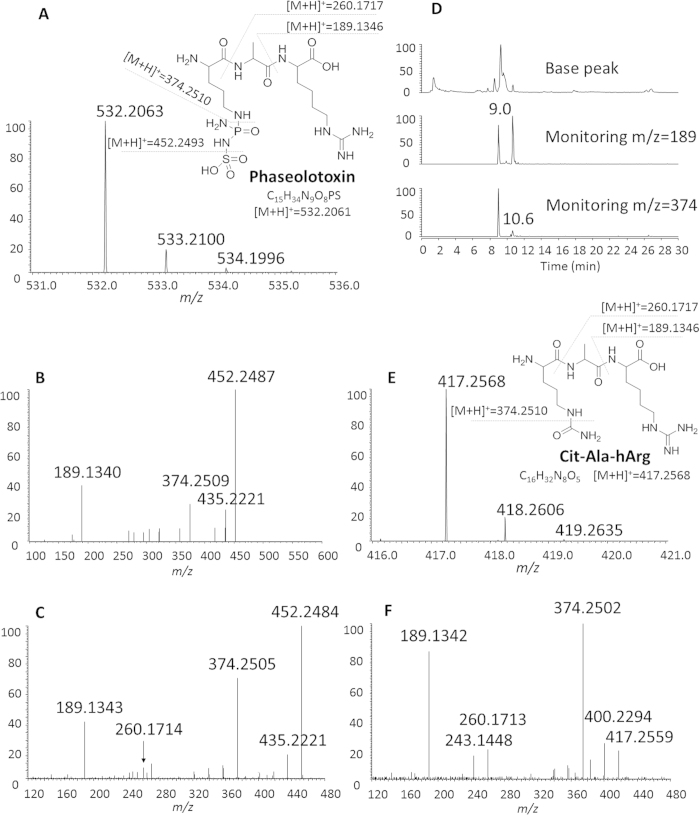
PHTs were detected in the cultural supernatants of wild type strain P. syringae pv. phaseolicola 1448A by High resolution mass spectrometry (HR MS) and 31P nuclear magnetic resonance (NMR) analysis. The 31P chemical shift of PHTs was 10.94 ppm (Figure S1). Tandem mass spectra (MS2) indicated that species with m/z ratio at 452, 374 and 189 are the characteristic daughter ions of PHTs (m/z ratio at 532) (Fig. 1B). Daughter ions with m/z ratio at 374 and 189 were also observed in the MS3 spectra when desulfonated PHTs (m/z ratio at 452) were further fragmented (Fig. 1C).
Figure 1. PIS mass spectrometric analysis to screen for PHT biosynthetic intermediates.
(A) high resolution mass spectrum of PHTs (inset is fragmentation pattern of PHT); (B) tandem mass (MS2) spectrum of PHTs; (C) tandem mass (MS3) spectrum of daughter ions with m/z ratio at 452.3 in panel (B); (D) chromatogram of PIS mass spectrometric analysis; (E) high resolution mass spectrum of novel ions with m/z ratio at 417.2568 (inset is fragmentation pattern of Cit-Ala-hArg); (F), tandem mass spectrum of ions with m/z ratio at 417.2568.
PHTs characteristic daughter ions with m/z ratio at 374 and 189 were used as queries to screen for PHT biosynthetic intermediates by PIS mass spectrometry. Novel ion with m/z ratio at 417 was detected and tandem mass spectra were further recorded on HR-FT-MS instrument (Fig. 1D,E). Ions with m/z ratio at 417, desulfonated PHTs and PHTs shared two identical daughter ions, m/z ratio at 374 and 189 (Fig. 1F), which are corresponding to the exact mass of tripeptides Orn-Ala-hArg and hArg residues respectively. It indicated that the novel ion with m/z ratio at 417 is a derivate of PHT peptide scaffold Orn-Ala-hArg. Based on exact mass of 417.2568 (Fig. 1E), the side functional group was deduced to be carbamoyl. Carbamylation of Orn-Ala-hArg produced Cit-Ala-hArg, which was a deduced chemical structure of the newly detected ions with m/z ratio at 417.
Tripeptides Cit-Ala-hArg were by-products of PHT biosynthetic pathway
To determine the biosynthetic relationship between tripeptides Cit-Ala-hArg and PHTs, P. syringae pv. phaseolicola 1448A genes phtU, phtQ and phtL were in-frame deleted (Figure S2). Neither Cit-Ala-hArg nor PHT was detected from the culture supernatants of phtU- mutant CL001 strain. Gene phtU complementation strain CL004 regained the ability to produce PHTs (Figure S3). Authentic L-Orn- L-Ala- L-hArg standards feeding restored phtU- mutants with the ability to produce PHTs at 18 °C, which is the temperature permissive for PHTs synthesis in wild type producers (Fig. 2B). Simultaneously, feeding of L-Cit- L-Ala- L-hArg and L-Ala- L-hArg restored phtU- mutants with the ability to produce PHTs as well (Figure S3). To the phtQ in-frame deletion, neither Cit-Ala-hArg nor PHT was detected from the culture supernatants of phtQ- mutant strain CL002. Surprisingly, PSOrn was detected from the culture supernatants of phtQ- mutants. HR tandem MS spectra of PSOrn were shown in Fig. 3. None of the authentic oligopeptides described here restored phtQ- mutants with the ability to produce PHTs (Fig. 2C). Meanwhile, gene phtQ complementation strain CL005 regained the ability to produce PHTs (Figure S3). Gene phtL knockout did not abolish phtL- mutant strain CL003 the ability to produce tripeptides Cit-Ala-hArg at 18 °C (Fig. 2D). PhtL is necessary for PHTs biosynthesis since phtL- mutants did not produce any PHT. The accumulated amounts of Cit-Ala-hArg were increased when PHT biosynthesis pathway was blocked in P. syringae pv. phaseolicola phtL- mutants (Fig. 2D).
Figure 2. Extracted ion chromatograms of compounds from culture supernatants of PHT producers and gene in-frame deletion mutants.
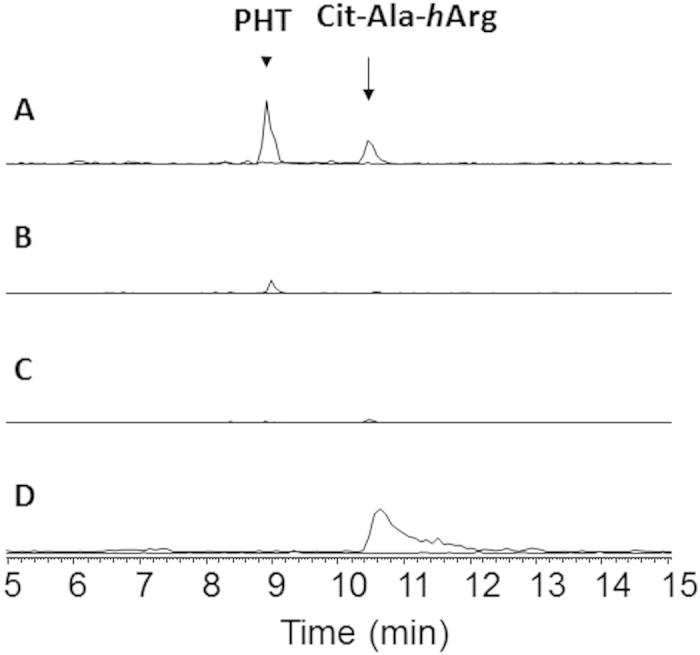
(A) 1448A (PHT producer, wild type); (B) phtU- mutant strain CL001 fed with Orn-Ala-hArg; (C) phtQ- mutant strain CL002 fed with Orn-Ala-hArg; (D) phtL- mutant. Upper lines were extracted for PHTs and lower lines were extracted for tripeptides Cit-Ala-hArg, with a tolerance of 0.5 Da.
Figure 3. HR MS spectra of PSOrn.
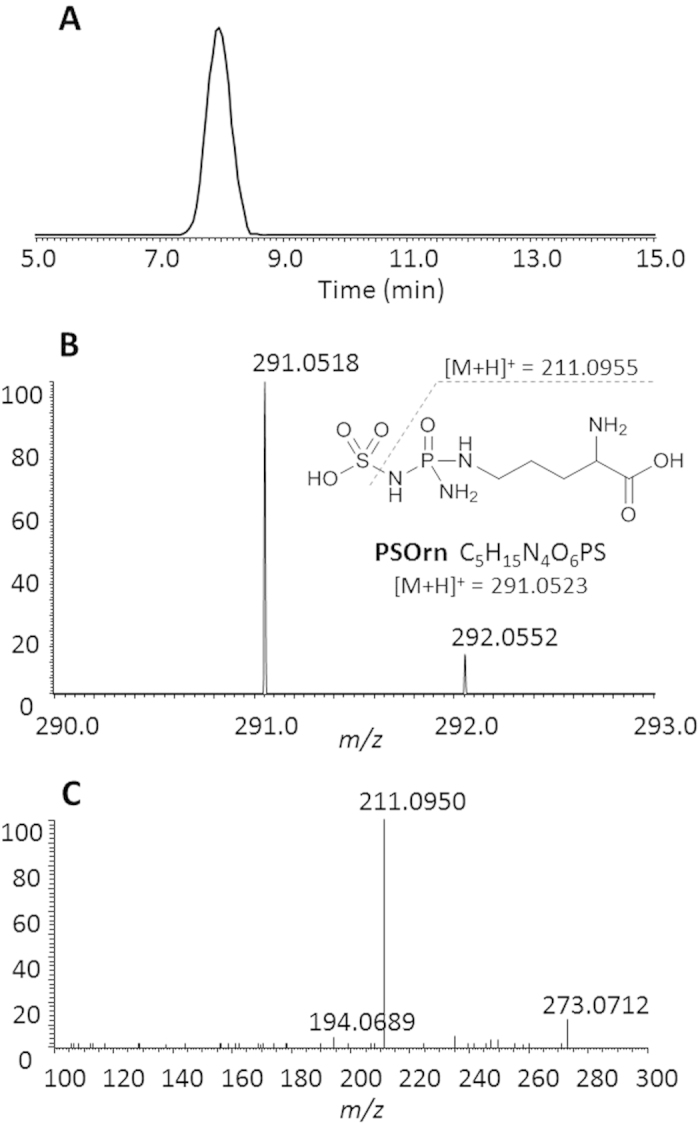
(A) Extracted ion chromatogram of PSOrn, with a tolerance of 5 ppm. (B) high resolution mass spectrum of PSOrn (inset is fragmentation pattern of PSOrn); (C) tandem mass (MS2) spectrum of PSOrn.
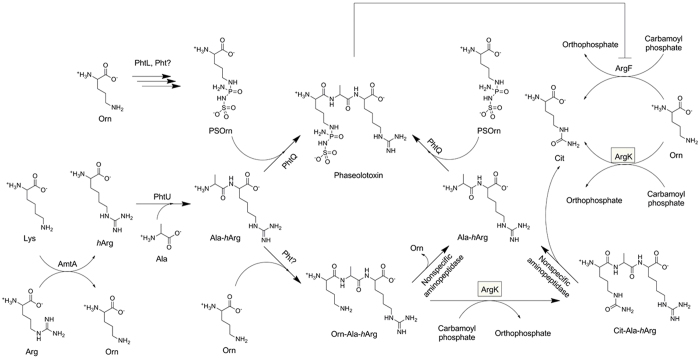
Gene deletion and feeding experiments results described here showed that tripeptides Cit-Ala-hArg were by-products accumulated during PHTs biosynthetic process. Dipeptide L-Ala- L-hArg was a joint precursor for the synthesis of both PHT and Cit-Ala-hArg (Fig. 4). Nonspecific aminopeptidase catalyzed hydrolysis of L-Orn- L-Ala- L-hArg and L-Cit- L-Ala- L-hArg would release L-Ala- L-hArg. Incorporation of L-Ala- L-hArg into PHT biosynthetic pathway restored phtU- mutants with PHTs producing ability (Fig. 4). Detection of precursor PSOrn from the culture supernatant of phtQ- showed that PhtQ involved in the biosynthesis of PHTs with PSOrn and L-Ala- L-hArg as substrates. Accumulation of by-products Orn-Ala-hArg and Cit-Ala-hArg from the culture supernatants of wild type strains and certain gene in frame deletion mutants might be attributed to the substrate tolerance of PhtQ. We can not rule out the possibility that there are certain uncharacterized peptide ligases participated in the formation of Orn-Ala-hArg and/or Cit-Ala-hArg. PhtL might be involved in the biosynthetic steps of PSOrn with L-Orn as a close precursor. Based on the fact that there is a phtL gene homologous (agnD1 and agnD2) in the biosynthetic gene cluster of another phosphoramidate natural product Agrocin 8428,29,30, we assume that phtL is related to the N-P bonds formation of PHTs.
Figure 4. Dual roles of ArgK in PHT producers self-defense.
ArgK provided alternative Arginine source by acting as a functional replacement of housekeeping OTCase ArgF, and also controlled the production of PHT by converting PHT biosynthetic precursors to nontoxic Cit-Ala-hArg reservoir.
ArgK catalyzed the formation of Tripeptides Cit-Ala-hArg
To extend our understanding about why such a great amount of by-products were accumulated, it is necessary to address the question concerning formation of tripeptides Cit-Ala-hArg. Two possible mechanisms could be involved to account for this phenomenon. First, PhtQ enzyme is a substrate tolerant peptide ligase and capable of catalyzing the synthesis of Cit-Ala-hArg with Citrulline and dipeptide Ala-hArg as substrates. Second, a potential transcarbamylase catalyzes the carbamylation of Orn-Ala-hArg to form Cit-Ala-hArg. To the PHTs producing Pseudomonas cells, it suffers from Citrulline shortage since the Citrulline formation reaction kcat catalyzed by ArgK reduced to between 1% and 2% of that catalyzed by typical OTCase, such as ArgF31. It is not an economical strategy to produce a great amount of by-product Cit-Ala-hArg by consuming limited Citrulline source. In addition, by-products Cit-Ala-hArg were detected from the cultural supernatants of phtU- as well as phtQ- mutants when Orn-Ala-hArg was added as substrate respectively (Figure S4). Therefore, the OTCase ArgK was suspected to be a main contributor for the formation of by-product Cit-Ala-hArg in vivo.
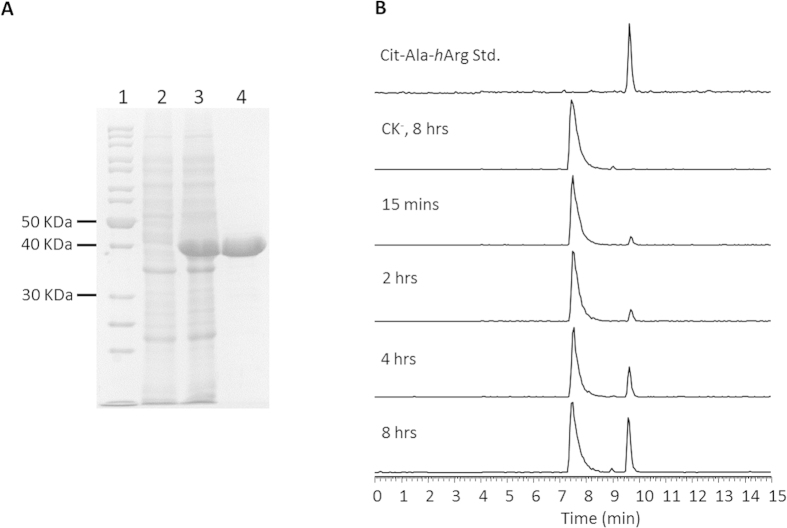
Enzyme assay showed that purified ArgK catalyzed the formation of L-Cit- L-Ala- L-hArg with L-Orn- L-Ala- L-hArg and carbamyl phosphates as substrates at 28 °C in vitro (Fig. 5). The tripeptides L-Cit- L-Ala- L-hArg production was positively correlated with reaction time, while no detectable amount was observed from the negative control reaction in which ArgK was absent (Fig. 5B). On the contrary, ArgK did not catalyze the reverse reaction in which the system was set up with L-Cit- L-Ala- L-hArg and orthophosphate as substrate.
Figure 5. PHT resistant OTCase ArgK enzyme assay.
(A) SDS-PAGE picture of purified ArgK protein. Lane 1, molecular weight marker; Lane 2, total protein without IPTG inducing; Lane 3, total protein with IPTG inducing; Lane 4, purified ArgK protein. (B) Total ion chromatograms of enzyme reaction system analyzed by HPLC-MS with a SIM mass spectrometric method screening m/z ratio from 360 to 430. Reaction system was set as 0.2 mg purified ArgK, 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1.5 mM Lithium carbamoyphosphate dibasic hydrate, 1 mM L-Orn- L-Ala- L-hArg, 2 mM MgSO4 and 0.5 mM ATP, 200 μL in total. CK- means negative control reaction in which ArgK was absent. Peaks of retention time at 7.5 min were substrates L-Orn- L-Ala- L-hArg; peaks of retention time at 9.6 min were products L-Cit- L-Ala- L-hArg.
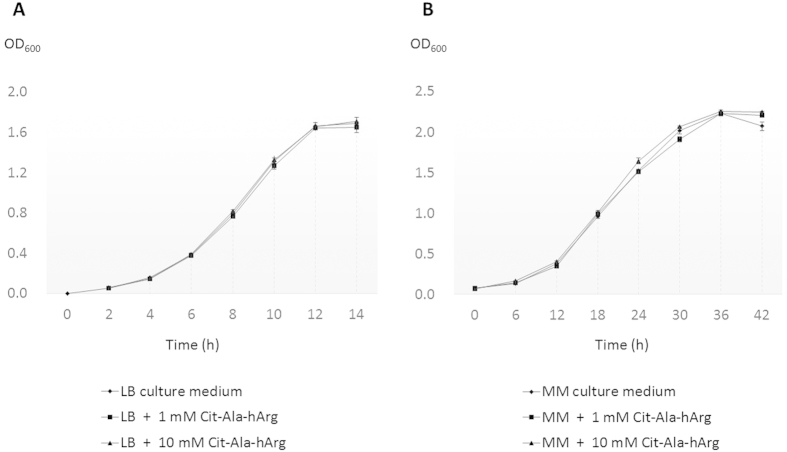
It indicated that ArgK played dual roles for PHT producers’ self-defense, which the first one was providing alternative Arginine source by acting as functional replacement of ArgF as documented previously13,14,15, and the second one was reducing PHTs production by modifying L-Orn- L-Ala- L-hArg to produce by-products L-Cit- L-Ala- L-hArg. Exogenous L-Cit- L-Ala- L-hArg with a concentration range from 1 to 10 mM did not affect the growth of PHTs producer P. syringae 1448A (Fig. 6). Tripeptides L-Cit- L-Ala- L-hArg were partially consumed by P. syringae cells in the tested conditions.
Figure 6. Growth curve of P. syringae pv. phaseolicola 1448A in distinct culture media.
(A) LB medium with additional 1 mM or 10 mM tripeptide L-Cit- L-Ala- L-hArg. (B) MM medium with additional 1 mM or 10 mM L-Cit- L-Ala- L-hArg.
Besides anabolic OTCase ArgF, another catabolic OTCase ArcB has been detected from P. aeruginosa32,33. ArcB is involved in the Arginine deiminase pathway and catalyzes the phosphorolysis of Citrulline to produce Ornithine and carbamyl phosphate34,35. It is exactly the reverse reaction of that one involved in the Arginine biosynthesis pathway catalyzed by ArgF. Phylogenetic analysis indicated that ArgK is more closely related to ArcB of P. aeruginosa PA01 than ArgF of P. syringae (Fig. 7). Comparing with typical anabolic OTCases, amino acid residue substitutions of ArgK were observed in the conserved sites around ornithine binding “SMG” loop31. The newly identified ArgK function announced here accords well with the alteration of substrate binding pocket. PHT resistant and substrate tolerant characters of OTCase ArgK challenge the current paradigm for OTCase.
Figure 7. Phylogenetic tree of OTCases analyzed by MEGA6 program.
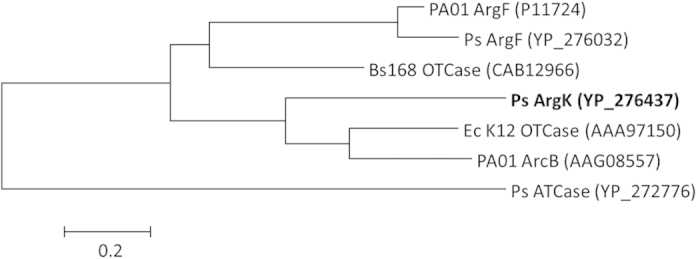
PA01 ArgF, OTCase ArgF of P. aeruginosa PA01; Ps ArgF, OTCase ArgF of P. syringae pv. phaseolicola 1448A; Bs168 OTCase, OTCase of Bacillus subtilis 168; Ps ArgK, PHT resistant OTCase ArgK of P. syringae pv. phaseolicola 1448A; Ec K12 OTCase, OTCase of Escherichia coli K-12; PA01 ArcB, catabolic OTCase ArcB of P. aeruginosa PA01; Ps ATCase, Aspartate carbamyltransferase of P. syringae pv. phaseolicola 1448A. Bracketed is the GenBank accession number.
Discussion
For pathogens colonization and subsequent symptom development, it is necessary to produce certain amount of virulence factors when P. syringae pv. phaseolicola cells infiltrated into plant tissues3,4. Phytotoxic compound PHTs conferred pathogens with survival advantages while burdened themselves with self-toxicity, amino acid source and energy expenditure simultaneously. The productivity and production of PHTs must be controlled accurately. Nevertheless, there is no typical transcriptional regulator gene has been identified from PHT biosynthetic gene cluster so far4,17.
To the PHT producers, it is a passive strategy to employ PHT resistant ArgK acting as a functional replacement of OTCase ArgF. In this study, we demonstrated that ArgK directly controlled the production of PHTs by carbamylation of Orn-Ala-hArg to produce by-product Cit-Ala-hArg. Cit-Ala-hArg could act as a nontoxic reservoir or be degraded by nonspecific aminopeptidase to provide additional Citrulline source (Fig. 4). It depends on the physiological situation of PHTs producers. This active manner could act as a complementary mechanism to the passive one for PHT producers’ self-defense. The dual roles of ArgK reported here help us to understand how P. syringae pv. phaseolicola kept a nice balance between amino acid fundamental metabolic pathway and the second metabolites PHTs biosynthesis pathway.
Materials and Methods
Fermentation for PHTs producing
Seed cells of P. syringae pv. phaseolicola strain 1448A and gene in-frame deletion mutants were cultured by LB medium at 28 °C. Cells were harvested and washed twice with equal volume of distilled water, then transferred into sucrose minimal medium and fermented at 18 °C for 72 hours for PHTs producing. In the feeding experiments, 0.5 mM authentic oligopeptides were fed in the fermentation process of phtU- and phtQ- mutants respectively.
PHT biosynthesis related genes in-frame deletion and complementation
PHTs native producer P. syringae pv. phaseolicola strain 1448A genome Fosmid library was constructed with CopyControl™ Fosmid Library Production Kit (Epicentre® Biotechnologies products) according to standard protocols36. Fosmid 7C6 with a 33.6 kb DNA insert which covers the complete PHT biosynthetic gene cluster and flanking sequence of 5.5 kb upstream and 3.3 kb downstream was screened out by PCR. Gene phtU of Fosmid 7C6 was replaced by Apramycin resistant gene aac(3)IV from plasmid pIJ773 by PCR targeting with primer pair DUF1/DUR2 (Table S1)37. Gene aac(3)IV was eliminated by FLP-recombinase mediated excision in E.coli DH5α/BT340 to generate a disruption cassette pWHU2001. Plasmids pWHU2001 were transferred into P. syringae pv. phaseolicola 1448A cells by electroporation and single crossover recombinant strains resistant to chloramphenicol were picked up. The phtU gene in-frame deletion mutant strains CL001 resulted from double crossover were selected out by rounds of relaxation and chloramphenicol sensitivity tests and then validated by PCR with primer pair VLF1/VLR2 (Figure S2A, S2B, Table S1). Identical strategy was employed for gene phtQ and phtL in-frame deletion. Primer pair DQF1/DQR2 (Table S1) was used in phtQ PCR targeting and primer pair VQF1/VQR2 (Table S1) was used in phtQ- mutant strain CL002 PCR validation (Figure S2C, S2D). Primer pair DLF1/DLR2 (Table S1) was used in phtL PCR targeting and primer pair VLF1/VLR2 (Table S1) was used in phtL- mutant strain CL003 PCR validation (Figure S2E, S2F).
To the gene complementation of phtU and phtQ in CL001 and CL002 strains, the constitutive promotor of gene phtA was employed to construct infusion genes PphtA-ORFphtU and PphtA-ORFphtQ. A 227 bp DNA fragment containing PphtA was amplified by PCR with primer pair PphtAF1/PphtAR2 (Table S1). The ORF region of gene phtU was amplified by PCR with primer pair PhtUF1/PhtUR2 (Table S1). These two fragments were insert into the BamHI site of a RK2-derived plasmid pRKaraRED to give recombinant plasmid pWHU2004. Transformation of pWHU2004 into phtU- mutant strain CL001 gave phtU gene complementation strain CL004. Identical strategy was employed for gene phtQ complementation strain CL005 construction with the exception of that phtQ was amplified with primer pair PhtQF1/PhtQR2 (Table S1) and the recombinant plasmid was defined as pWHU2005.
ArgK over-expression and enzyme assay
Gene argK was amplified by PCR with primer pair OKF1/OKR2 (Table S1) and inserted into the BamHI/XhoI locus of vector pET28a. His·tag-ArgK fusion proteins were over-expressed and purified with standard protocol36. Authentic standards of L-Orn- L-Ala- L-hArg and L-Cit- L-Ala- L-hArg were purchased from Life Tein L.T.D. Company (Beijing) (Figure S3). Transcarbamylase enzyme assay were performed in 1.5 mL eppendorf tubes with a system as 0.2 mg purified ArgK, 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1.5 mM Lithium carbamoyphosphate dibasic hydrate, 1 mM L-Orn- L-Ala- L-hArg, 2 mM MgSO4 and 0.5 mM ATP, 200 μL in total. ArgK was absent in the negative control reactions while the other factors remained identical. The enzyme was removed from the reaction system with a 10 KDa centrifugal filter at 10,000 rpm for 10 minutes. The residual sections were diluted to ten times volume of the original reaction system before injected into the HPLC-MS analyzer.
HPLC-MS analysis
Culture supernatants for PHTs and biosynthetic intermediates discovery were thirty times concentrated by rotovaping at room temperature and then cut by methanol with a final concentration of 90%. Samples were dried out by rotovaping and re-suspended in ddH2O for HPLC-MS analysis. A Thermo hypercarb column (100 × 2.1 mm, 5 μm, column temperature 30 °C) with a flow rate of 0.4 mL/min was employed in the HPLC section. The HPLC gradient was 0% solvent B (4 min), 0–40% solvent B (12 min), 40–100% solvent B (3 min), 100% solvent B (3 min), 100–0% solvent B (3 min), 0% solvent B (5 min). Solvent A was 0.1% formic acid in H2O and solvent B was 0.1% formic acid in CH3CN.
High resolution MS analysis was carried out in a positive mode on a Thermo Scientific LTQ XL Orbitrap mass spectrometer as described previously38. PHTs samples from the culture supernatants of P. syringae pv. phaseolicola strain 1448A were used to build the MS fragmentation fingerprint of PHTs and desulfonated PHTs by selected ion reaction (SIR) method. Daughter ions with m/z ratio at 189 and 374 were recognized to be the characteristic fragments of PHTs. To screen for PHTs biosynthetic intermediates, PIS was performed in a positive mode on a Thermo Scientific TSQ Quantum Access MAX instrument (monitoring m/z ratio at 189 and 374) equipped with a Thermo Scientific Accela 600 pump. Target ions were selected out for HR tandem MS analysis as described above to figure out the elemental compositions.
Enzyme assay samples were analyzed on the TSQ instrument with a selected ion monitoring (SIM) scan mode with a range from m/z ratio at 360 to 430. Reaction products were double checked by HR tandem MS analysis on the Orbitrap instrument.
NMR analysis
1H NMR and 13C NMR spectra of tripeptides L-Orn- L-Ala- L-hArg (20 mg) and L-Cit- L-Ala- L-hArg (20 mg) were recorded on Agilent 400 MHz instrument in D2O, respectively (Figure S3). Culture supernatants of P. syringae pv. phaseolicola strain 1448A were cut by methanol and dried out as described above. Residues were re-suspended in ddH2O with 10% D2O and 31P NMR spectra were recorded on Agilent 400 MHz instrument.
Bioassay of P. syringae 1448A against L-Cit- L-Ala- L-hArg
To evaluate the toxicity of L-Cit- L-Ala- L-hArg to PHT producers P. syringae 1448A, authentic tripeptide L-Cit- L-Ala- L-hArg was added to the culture media of 1448A. Two kinds of media, LB (Tryptone 10.0 g, Yeast extract 5.0 g and NaCl 5.0 g for 1 liter, pH 7.2) and MM (Sucrose 5.0 g, KH2PO4 2.0 g, (NH4)2HPO4 2.0 g, MgSO4·7H2O 1.0 g, FeSO4·7H2O 0.01 g and MnSO4 0.01 g for 1 liter, pH 7.2), were employed in the assay. Two kinds of compound concentrations, 1 mM and 10 mM, were tested respectively. The OD600 value of bacterial cultures at different time points was monitored by a Nanodrop 2000 spectrophotometer.
Additional Information
How to cite this article: chen, L. et al. Ornithine Transcarbamylase ArgK Plays a Dual role for the Self-defense of Phaseolotoxin Producing Pseudomonas syringae pv. phaseolicola. Sci. Rep. 5, 12892; doi: 10.1038/srep12892 (2015).
Supplementary Material
Acknowledgments
The authors greatly appreciate Dr. Prof. Jesús Murillo from Universidad Pu´blica de Navarra, Spain for the gift of PHTs producer Pseudomonas syringae pv. phaseolicola 1448A. The authors would like to acknowledge financial support from Ministry of Science and Technology of China (“973” Program 2013CB734003 and “863” Program 2012AA02A701) and National Natural Science Foundation of China (81273411 and 31200037).
Footnotes
Author Contributions Conceived and designed the experiments: Z.D. and C.Z. Performed the experiments: L.C. and P.L. Analyzed the data: L.C. and C.Z. Contributed reagents/materials/analysis tools: L.C. and P.L. Wrote the paper: L.C. and C.Z.
References
- Hirano S. S. & Upper C. D. Population Biology and Epidemiology of Pseudomonas Syringae. Annu Rev Phytopathol 28, 155–177 (1990). [Google Scholar]
- Hirano S. S. & Upper C. D. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev 64, 624–653 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C. L., Alarcon-Chaidez F. & Gross D. C. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev 63, 266–292 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross H. & Loper J. E. Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep 26, 1408–1446 (2009). [DOI] [PubMed] [Google Scholar]
- Renzi M. et al. Bacterial canker on kiwifruit in Italy: anatomical changes in the wood and in the primary infection sites. Phytopathology 102, 827–840 (2012). [DOI] [PubMed] [Google Scholar]
- Langley D. B., Templeton M. D., Fields B. A., Mitchell R. E. & Collyer C. A. Mechanism of inactivation of ornithine transcarbamoylase by Ndelta -(N’-Sulfodiaminophosphinyl)-L-ornithine, a true transition state analogue? Crystal structure and implications for catalytic mechanism. J Biol Chem 275, 20012–20019 (2000). [DOI] [PubMed] [Google Scholar]
- Mitchell R. E. & Bieleski R. L. Involvement of phaseolotoxin in halo blight of beans: transport and conversion to functional toxin. Plant Physiol 60, 723–729 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. E., Niemczura W. P., Kwok O. C. H. & Patil S. S. Inhibitors of Ornithine Carbamoyltransferase from Pseudomonas syringae pv. phaseolicola. Revised Structure of Phaseolotoxin. Tetrahedron Lett 25, 3931–3934 (1984). [Google Scholar]
- Turner J. G. Effect of phaseolotoxin on the synthesis of arginine and protein. Plant Physiol 80, 760–765 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell R. E., Johnston J. S. & Ferguson A. R. Phaseolotoxin and other phosphosulphanyl compounds: biological effects. Physiol Plant Pathol 19, 227–235 (1981). [Google Scholar]
- Staskawicz B. J. & Panopoulos N. J. A rapid and sensitive microbiological assay for phaseolotoxin. Phytopathology 69, 663–666 (1979). [Google Scholar]
- Templeton M. D., Sullivan P. A. & Shepherd M. G. The inhibition of ornithine transcarbamoylase from Escherichia coli W by phaseolotoxin. Biochem J 224, 379–388 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosqueda G., Van den Broeck G., Saucedo O., Bailey A. M., Alvarez-Morales A. & Herrera-Estrella L. Isolation and characterization of the gene from Pseudomonas syringae pv. phaseolicola encoding the phaseolotoxin-insensitive ornithine carbamoyltransferase. Mol Gen Genet 222, 461–466 (1990). [DOI] [PubMed] [Google Scholar]
- Peet R. C. & Panopoulos N. J. Ornithine carbamoyltransferase genes and phaseolotoxin immunity in Pseudomonas syringae pv. phaseolicola. EMBO J 6, 3585–3591 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz B. J., Panopoulos N. J. & Hoogenraad N. J. Phaseolotoxin-insensitive ornithine carbamoyltransferase of Pseudomonas syringae pv. phaseolicola: basis for immunity to phaseolotoxin. J Bacteriol 142, 720–723 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrebola E., Cazorla F. M., Perez-Garcia A. & de Vicente A. Chemical and metabolic aspects of antimetabolite toxins produced by Pseudomonas syringae pathovars. Toxins (Basel) 3, 1089–1110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera S. et al. Functional characterization of the gene cluster from Pseudomonas syringae pv. phaseolicola NPS3121 involved in synthesis of phaseolotoxin. J Bacteriol 189, 2834–2843 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genka H. et al. Comparative analysis of argK-tox clusters and their flanking regions in phaseolotoxin-producing Pseudomonas syringae pathovars. J Mol Evol 63, 401–414 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang Y., Rowley K. B. & Patil S. S. Genetic organization of a cluster of genes involved in the production of phaseolotoxin, a toxin produced by Pseudomonas syringae pv. phaseolicola. J Bacteriol 175, 6451–6458 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuske J. & Fritsche W. Phaseolotoxin production by Pseudomonas syringae pv. phaseolicola: the influence of temperature. J Basic Microbiol 29, 441–447 (1989). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. Role of phaseolotoxin production by Pseudomonas syringae pv. actinidiae in the formation of halo lesions of kiwifruit canker disease. Physiol Mol Plant Pathol 60, 207–214 (2002). [Google Scholar]
- Aguilera S., De la Torre-Zavala S., Hernandez-Flores J. L., Murillo J., Bravo J. & Alvarez-Morales A. Expression of the gene for resistance to phaseolotoxin (argK) depends on the activity of genes phtABC in Pseudomonas syringae pv. phaseolicola. PLoS One 7, e46815 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lopez K., Hernandez-Flores J. L., Cruz-Aguilar M. & Alvarez-Morales A. In Pseudomonas syringae pv. phaseolicola, expression of the argK gene, encoding the phaseolotoxin-resistant ornithine carbamoyltransferase, is regulated indirectly by temperature and directly by a precursor resembling carbamoylphosphate. J Bacteriol 186, 146–153 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley K. B., Xu R. & Patil S. S. Molecular analysis of thermoregulation of phaseolotoxin-resistant ornithine carbamoyltransferase (argK) from Pseudomonas syringae pv. phaseolicola. Mol Plant Microbe Interact 13, 1071–1080 (2000). [DOI] [PubMed] [Google Scholar]
- Gonzalez-Villanueva L., Arvizu-Gomez J. L., Hernandez-Morales A., Aguilera-Aguirre S. & Alvarez-Morales A. The PhtL protein of Pseudomonas syringae pv. phaseolicola NPS3121 affects the expression of both phaseolotoxin cluster (Pht) and Non-Pht encoded genes. Microbiol Res 169, 221–231 (2014). [DOI] [PubMed] [Google Scholar]
- Hernandez-Guzman G. & Alvarez-Morales A. Isolation and characterization of the gene coding for the amidinotransferase involved in the biosynthesis of phaseolotoxin in Pseudomonas syringae pv. phaseolicola. Mol Plant Microbe Interact 14, 545–554 (2001). [DOI] [PubMed] [Google Scholar]
- Arai T. & Kino K. A novel L-amino acid ligase is encoded by a gene in the phaseolotoxin biosynthetic gene cluster from Pseudomonas syringae pv. phaseolicola 1448A. Biosci Biotechnol Biochem 72, 3048–3050 (2008). [DOI] [PubMed] [Google Scholar]
- Kim J. G. et al. Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the Trojan horse antibiotic that controls crown gall. Proc Natl Acad Sci USA 103, 8846–8851 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra S. et al. Plant tumour biocontrol agent employs a tRNA-dependent mechanism to inhibit leucyl-tRNA synthetase. Nat Commun 4, 1417 (2013). [DOI] [PubMed] [Google Scholar]
- Reader J. S. et al. Major biocontrol of plant tumors targets tRNA synthetase. Science 309, 1533 (2005). [DOI] [PubMed] [Google Scholar]
- Templeton M. D., Reinhardt L. A., Collyer C. A., Mitchell R. E. & Cleland W. W. Kinetic analysis of the L-ornithine transcarbamoylase from Pseudomonas savastanoi pv. phaseolicola that is resistant to the transition state analogue (R)-N delta-(N’-sulfodiaminophosphinyl)-L-ornithine. Biochemistry 44, 4408–4415 (2005). [DOI] [PubMed] [Google Scholar]
- Itoh Y. et al. Anabolic ornithine carbamoyltransferase of Pseudomonas aeruginosa: nucleotide sequence and transcriptional control of the argF structural gene. J Bacteriol 170, 2725–2734 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Wauven C., Pierard A., Kley-Raymann M. & Haas D. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J Bacteriol 160, 928–934 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeret V., Tricot C., Stalon V. & Dideberg O. Crystal structure of Pseudomonas aeruginosa catabolic ornithine transcarbamoylase at 3.0-A resolution: a different oligomeric organization in the transcarbamoylase family. Proc Natl Acad Sci USA 92, 10762–10766 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha Y., McCann M. T., Tuchman M. & Allewell N. M. Substrate-induced conformational change in a trimeric ornithine transcarbamoylase. Proc Natl Acad Sci USA 94, 9550–9555 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. F. & Russell D. W. Molecular Cloning: A Laboratory Manual, 3rd ed. Cold Spring Harbor Laboratory Press (2001). [Google Scholar]
- Bibb M. J., Buttner M. J., Chater K. F. & Hopwood D. A. Practical Streptomyces Genetics. John Innes Foundation (2000). [Google Scholar]
- Hao C., Huang S., Deng Z., Zhao C. & Yu Y. Mining of the pyrrolamide antibiotics analogs in Streptomyces netropsis reveals the amidohydrolase-dependent “iterative strategy” underlying the pyrrole polymerization. PLoS One 9, e99077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.