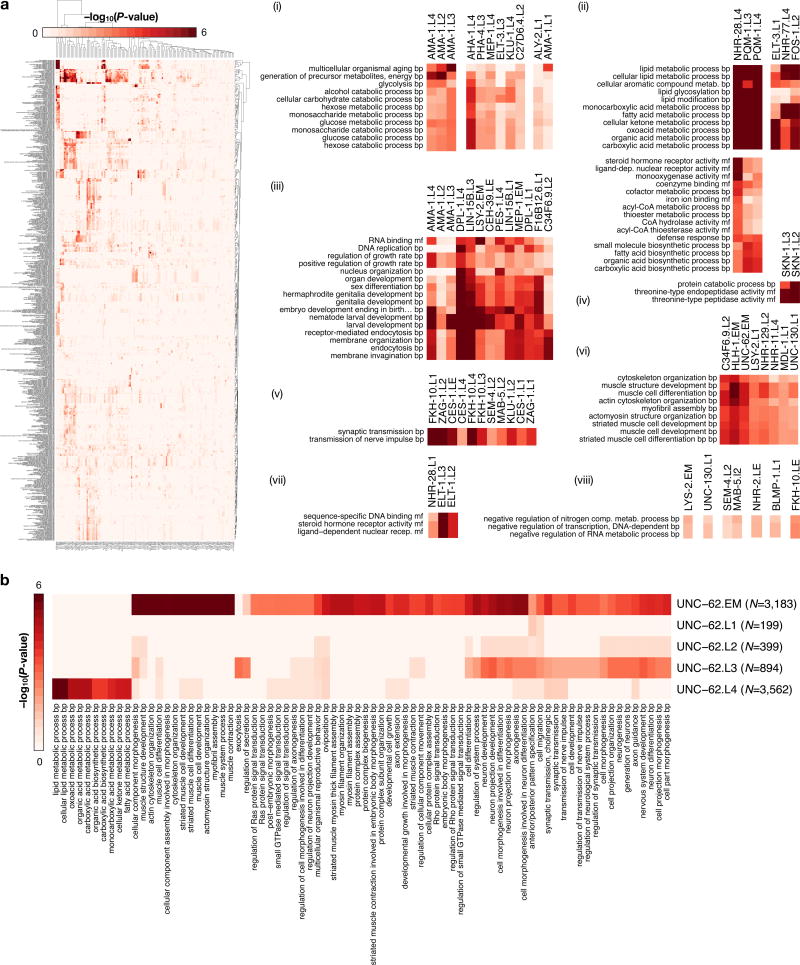
Extended Data Figure 4.
(a) Gene ontology (GO) enrichment matrix for 150 binding experiments (75 factors) spanning 6,347 significant GO enrichments (BH-corrected, P < 0.05) across 713 GO terms (level ≥4). For each experiment, GO-term enrichment was performed on gene targets as defined by binding within 1 kb of TSSs (ChipPeakAnno) 31. Enrichments for biological process (BP) and molecular function (MF) ontologies are shown, with distinct sets of enrichments highlighted (i–viii). (b) GO term enrichments among targets of UNC-62 binding show dramatical changes in the functional role of UNC-62 regulatory activity through development. Biological process (BP) terms (levels ≥4) enriched in UNC-62 libraries are shown. The number of UNC-62 binding sites identified per stage is indicated in parenthesis. Although changes in targets between mid-larval and adult stages have been suggested previously22, our analyses (performed with uniformly called binding sites) and expanded data indicate that the most dramatic changes occur between embryo and L4 larval stages. (+) MEP-1 indicates experiments performed in strain OP102.