Fig 5. Analysis of Piwi genes identified in C.hongkongensis transcriptome.
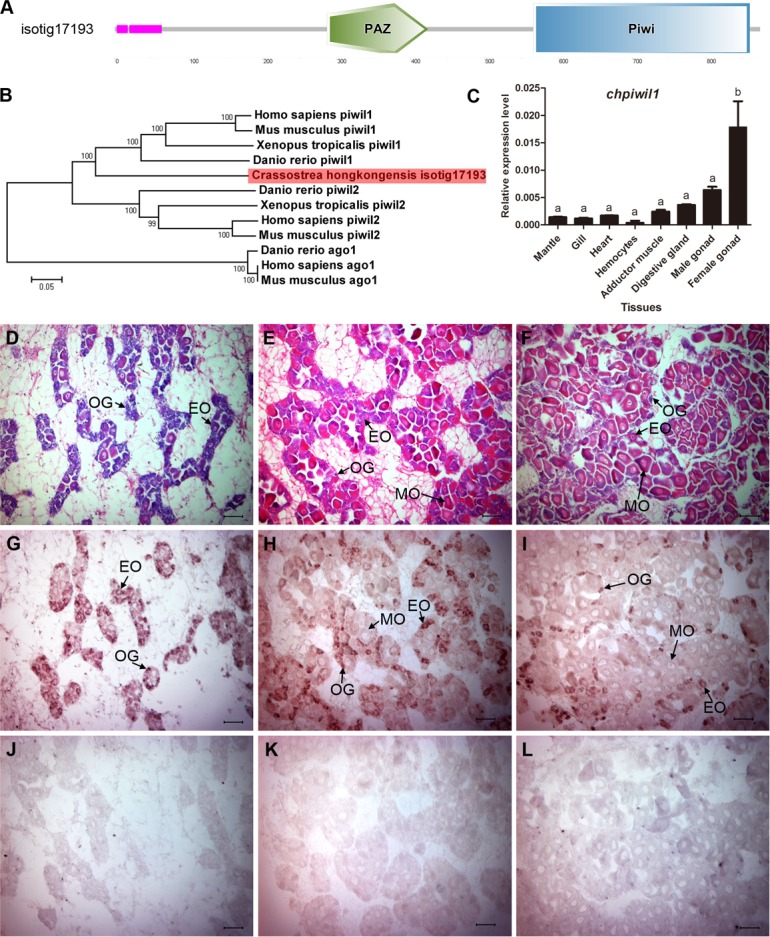
(A) Domain structure of the C.hongkongensis Piwi orthologs. (B) Phylogenetic tree constructed using the neighbor-joining method on the basis of the amino acid sequences alignment of C.hongkongensis Piwi ortholog with Piwi-like proteins of vertebrates. A total of 1000 bootstrap trials were run. Bootstrap values were indicated at each branch node. The scale bar represents an evolutionary distance of 0.1 amino acid substitutions per position. The transcript encoding C.hongkongensis Piwi ortholog, colored by red, was aligned to the clade of vertebrate Piwil1proteins. For the GenBank accession numbers of the reference sequences, see S3 Table. (C) Expression profile of ChPiwil1 in adult tissues by qRT-PCR with mean±s.e.m. as error bars (n = 5). (D-L) Expression profile of ChPiwil1during C.hongkongensis oogenesis by ISH. Histological analysis of C.hongkongensis female gonad at early developing stage (D), late developing stage (E) and mature stage (F) stained with HE. Expression profile of ChPiwil1during C.hongkongensis oogenesis by ISH with antisense (G-I) and sense probe (J-L). Positive cells are stained with brown or purple. OG: oogonia; EO: early vitellogenic oocyte; MO: mature ova.
