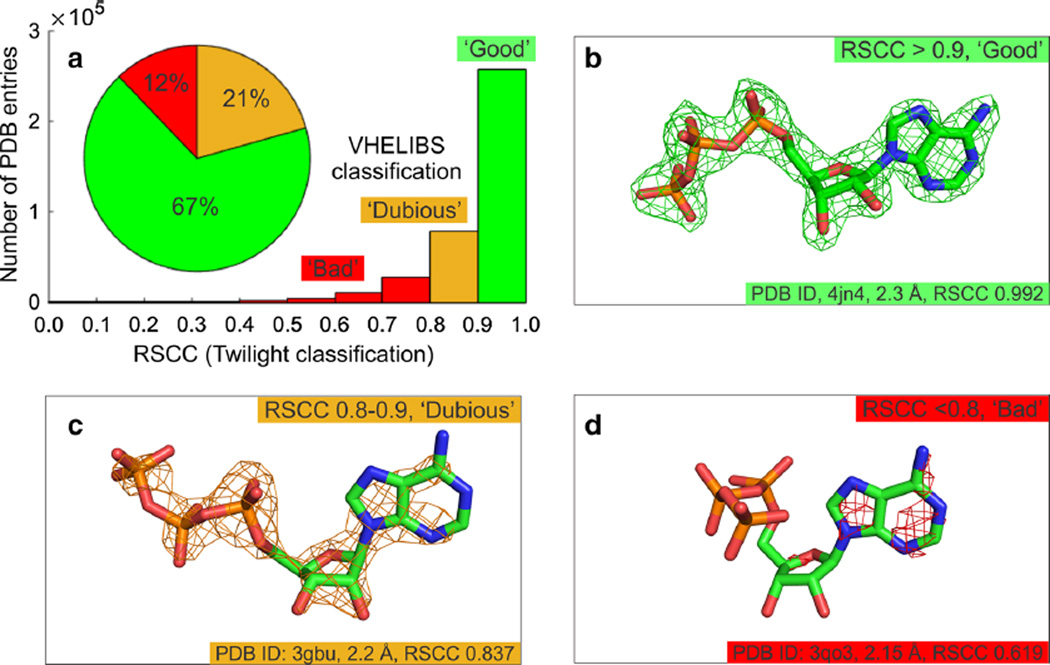
Fig. 3.
Electron density-based validation of protein–ligand models a Plots showing the distribution of the fits of ligands to their experimental electron density. Classifications were determined using Twilight [12, 15] and VHELIBS [14]. b through d demonstrate the fit of the ligand to the experimental electron density for a series of protein–ligand models of adenosine triphosphate (ATP, shown as sticks). Carbon atoms are shown in green, nitrogen in blue, oxygen in red and phosphorus in orange. The σA-weighted 2mFo–DFc electron density map is shown as a mesh contoured at 2σ. Data are shown in Table 1