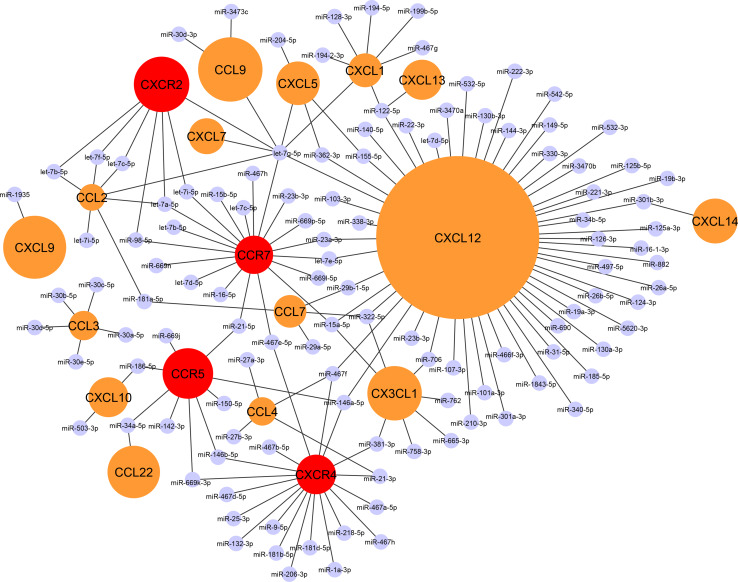
Fig. 1.
The chemokine–miRNA interactome: a classification of chemokines according to their miRNA-regulation pattern. Black lines are indicating experimentally validated interactions of miRNAs within the 3′ UTR region of murine chemokine or chemokine receptor transcripts using the web tool DIANA-TarBase v7.0 [109]. Chemokines/chemokine receptors without validated miRNA interactions have not been included in the interactome. The size of the circles in orange (chemokines) or red (chemokine receptors) corresponds to the number of predicted miRNA-binding sites in the 3′ UTR of the respective transcripts. A substantial number of putative miRNA-binding sites is predicted for CXCL12, which is in accordance with a high number of experimentally validated miRNA–CXCL12 interactions. In fact, a striking majority (53 %) of all validated miRNA interactions among all chemokines were observed for CXCL12 alone. In contrast, a limited number of miRNA interactions is predicted for other chemokines such as CCL2 or CXCL1 and for chemokine receptors, in accordance with a low number of functionally validated miRNA-binding sites for those transcripts