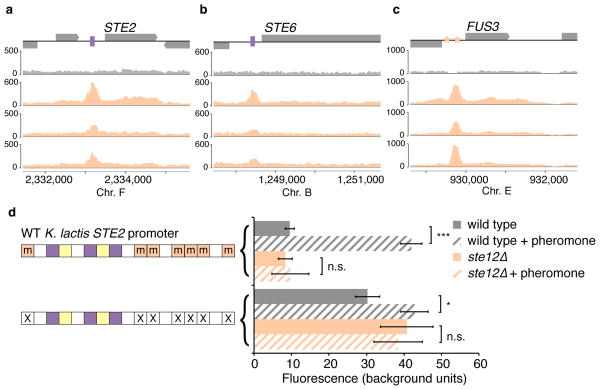
Figure 2. Ste12 is recruited to asg promoters in K. lactis without its binding site.
a–c, Chromatin immunoprecipitation of Ste12-myc using anti-myc antibodies (orange tracks) and an untagged control (gray track). The y-axis indicates sequencing read coverage. Matches to the Ste12 binding site are shown as orange stars, and matches to the a2-Mcm1 binding site are shown as pink rectangles. Shown are two conserved a-specific genes (a, b), one general pheromone-activated genes (c). d, At left, organization of the K. lactis STE2 regulatory region. cis-regulatory sites for a2, Mcm1, and Ste12 are shown in pink, green, and orange, respectively. Sites marked with an “m” reflect DNA sequences that contain a one-basepair mismatch to the 7-basepair consensus Ste12 cis-regulatory site. At right, the K. lactis STE2 regulatory region was fused to GFP and fluorescence was measured by flow cytometry. Shown is the mean fluorescence of three independent genetic isolates +/− s.d. Bottom, the construct contains two point mutations in each of the mismatched Ste12 cis-regulatory sites that should destroy any residual Ste12 binding to these sites. Although this construct is pheromone-inducible, we note that the basal expression of the K. lactis promoter increased when the mismatched Ste12 sites were mutated, probably through the creation of a binding site for another activator. One asterisk, P < 0.05, three asterisks, P < 0.001, by ANOVA followed by Tukey’s honest significant difference; for clarity, only the most relevant relationships are shown.