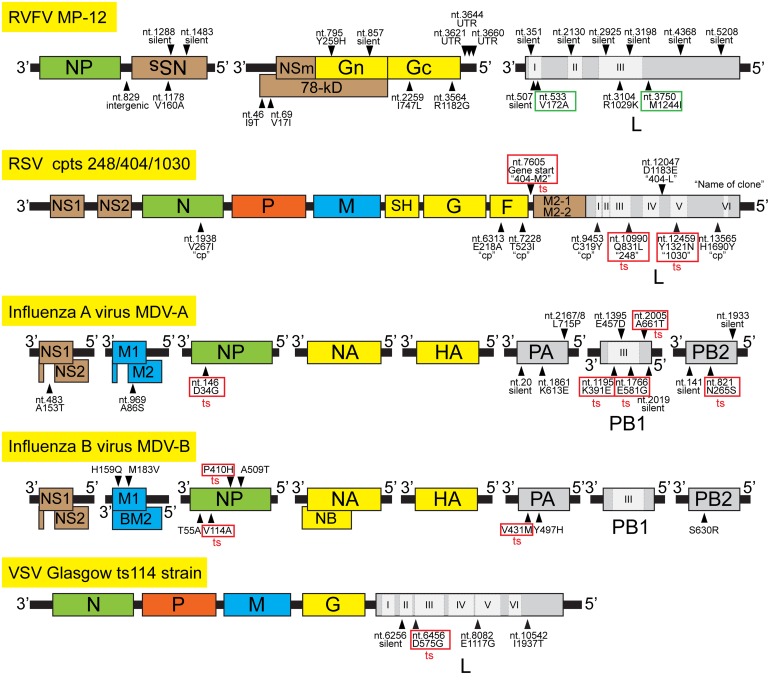
FIGURE 1.
Mapping of temperature-sensitive (ts) mutations for selected negative-stranded RNA viruses. The genome structures of RVFV MP-12 strain, Respiratory syncytial virus (RSV) cpts 248/404/1030, Influenza A virus (MDV-A), Influenza B virus (MDV-B), and Vesicular stomatitis virus (VSV) ts114 strain are shown. Viral genes are shown in different colors: Nucleocapsid protein (green), phosphoprotein (orange), RNA-dependent RNA polymerase (gray), envelope protein (yellow), matrix proteins (blue), and accessory or non-structural proteins (brown). Conserved six functional regions of RNA-dependent RNA polymerase (RdRp) among non-segmented negative-stranded RNA viruses are shown as I, II, III, IV, V, and VI, and those aligned to RdRp of bunyavirus and influenza virus are also indicated. Location of representative mutations and ts mutations (red square) are indicated by arrowheads. Putative ts mutations in the L-segment of MP-12 are shown in green squares. For RSV mutations, the name of clone, which encodes the mutation, is also shown in quotation.