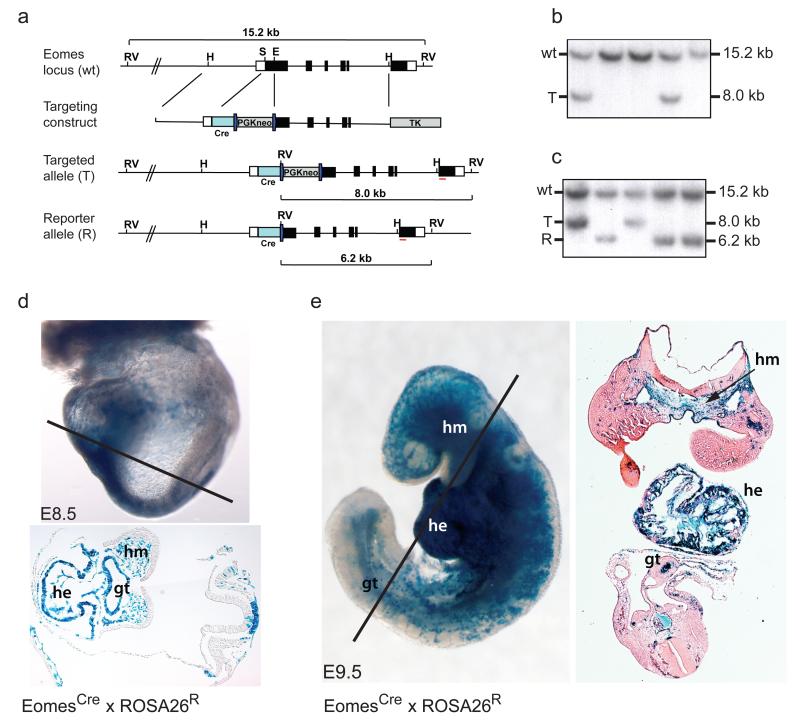
Figure 1.
Fate mapping EomesCre expressing cells reveals selective contributions to DE and cardiovascular cell lineages.
(a) Targeting strategy used to generate the EomesCre reporter allele. Cre recombinase coding sequences were inserted into Exon1 of the Eomes locus. RV, EcoRV; H, HpaI; S, SphI; E, EagI; Cre, Cre recombinase. (b) ES clones were screened by Southern blot on EcoRV digested DNA using a 3′ probe (red line in a) to detect wildtype (wt) and targeted (T) alleles. (c) Correctly targeted ES cells were transiently transfected with Cre to excise the PGK-neo selection cassette and generate the reporter allele (R), as shown by Southern blot. (d, e) Fate mapping experiments demonstrate that descendants of EomesCre expressing cells contribute to the myocardium and endocardium of the heart (he), the head mesenchyme (hm), vasculature and the endoderm of the primary gut tube (gt), but rarely colonize other mesoderm tissues formed from paraxial and lateral plate mesoderm. Sections were counterstained with eosin to highlight non-labelled cells.