Figure 1.
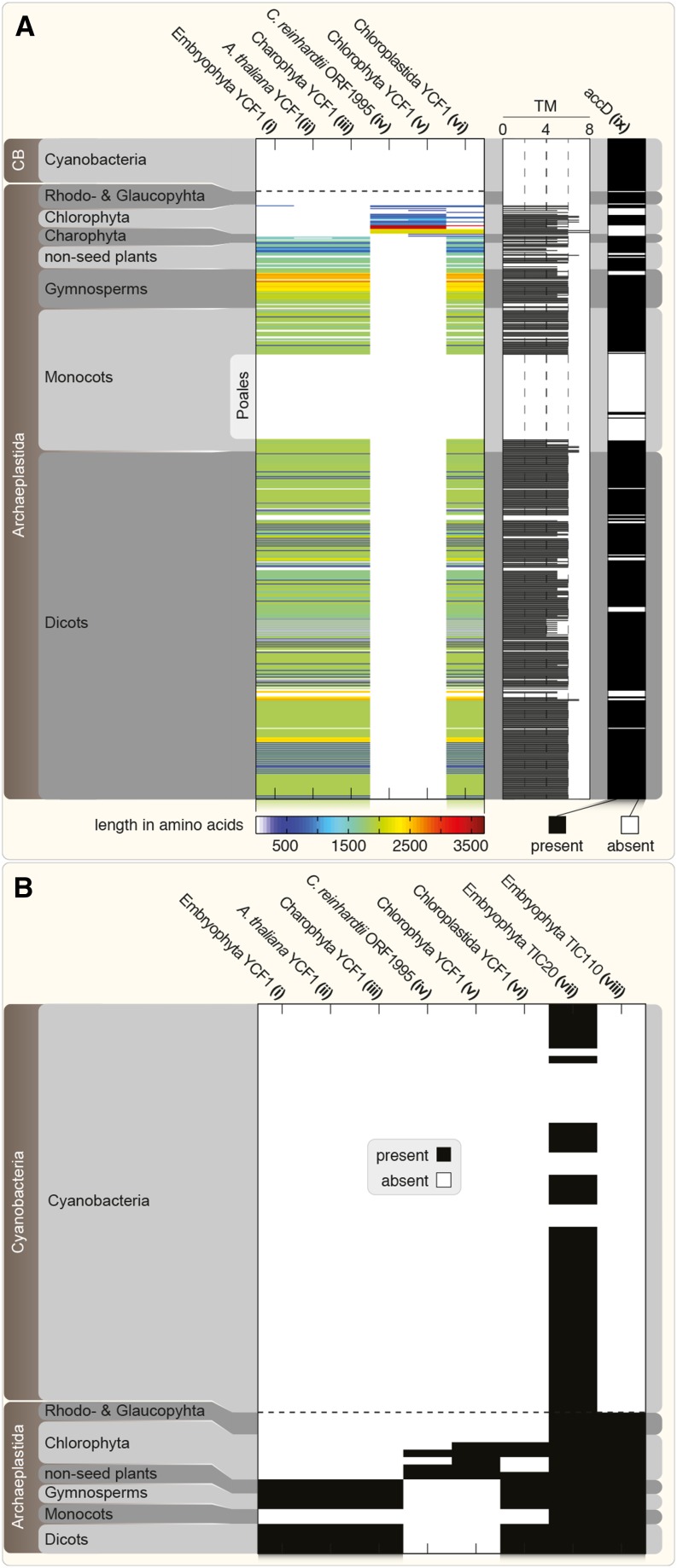
ycf1 Distribution and Sequence Diversity.
(A) Phylogenetic distribution of YCF1 among 558 archaeplastidal plastid and 55 cyanobacterial genomes. HMM searches were performed based on different alignments or sequence seeds (i to viii). Hits (E-value < 10−5) based on YCF1 alignments (i to vi) among all protein-encoding genes of 558 archaeplastidal plastid and cyanobacterial (CB) genomes are shown on the left, color-coded based on their amino acid length (blue to red; white means absence). Note the absence of YCF1 homologs among cyanobacteria, Rhodophyta, Glaucophyta, and Poales. For all detected YCF1 homologs, the number of TMHMM-predicted transmembrane helices (TM) is shown in the horizontal histogram. For comparison, the results (positive hit, black; no hit, white) of an HMM search against all plastid and cyanobacterial genomes using an alignment of 37 ACCD protein sequences is shown on the right (ix).
(B) The presence or absence (black and white, respectively) of genes encoding TIC20 and TIC110 among all protein-encoding genes of archaeplastidal nuclear and cyanobacterial genomes is shown. The screen is based on HMM searches using embryophyte sequence alignments (vii and viii). In comparison, the data from the YCF1 HMM searches in the archaeplastidal plastid and cyanobacterial genomes are shown in the same fashion. Dashed horizontal lines separate the cyanobacterial and archaeplastidal genome data. HMM and TMHMM results for every species analyzed are individually listed in Supplemental Data Set 1.