Figure 5.
SWP73A and SWP73B Differently Affect Flower Development and Flowering Time Control.
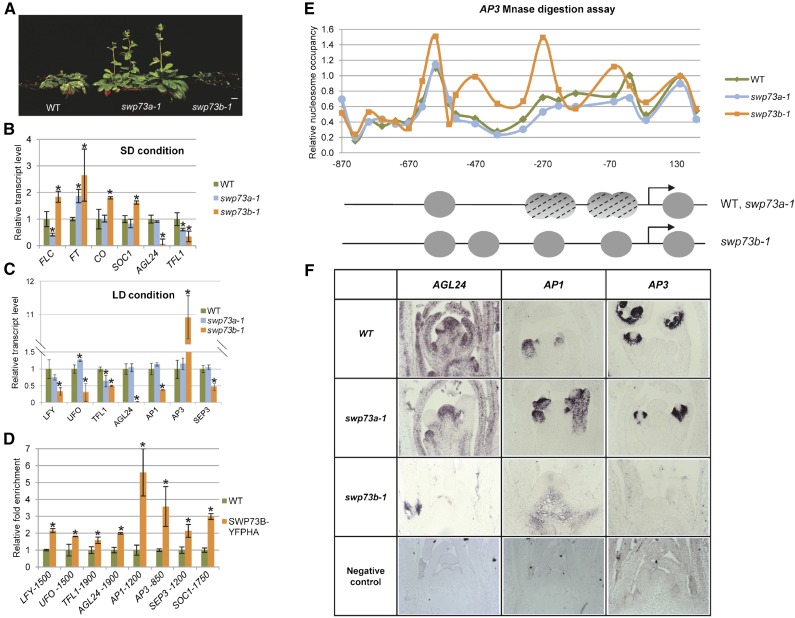
(A) Comparison of 8-week-old SD-grown wild-type, swp73a-1, and swp73b-1 plants. Bar = 1 cm.
(B) qRT-PCR analysis of expression of genes involved in control of flowering in 4-week-old SD-grown plants. Asterisks indicate significant differences from wild-type plants (error bars refer to sd, P < 0.05, Student’s t test, three biological and three technical replicates were used).
(C) qRT-PCR analysis of expression of genes involved in the control of floral meristem identity in 2-week-old LD-grown plants. Asterisks indicate significant differences from wild-type plants (error bars refer to sd, P < 0.05, Student’s t test, three biological and three technical replicates were used).
(D) ChIP-qPCR analysis of SWP73B location in the promoter regions of LFY, UFO, TFL1, AGL24, AP1, AP3, SEP3, and SOC1 genes in wild-type and SWP73B-YFPHA plants using the TA3 retrotransposon as reference. Asterisks indicate significant differences from wild-type plants (error bars refer to sd, P < 0.05, Student’s t test, three biological and three technical replicates were used).
(E) Relative nucleosome occupancy of AP3 promoter in 2-week-old wild-type, swp73a-1, and swp73b-1 plants. Upper panel: Nucleosome occupancy of AP3 promoter analyzed by MNase digestion followed by qPCR with tiled primers. The fraction of undigested genomic DNA amplified for each amplicon was normalized to that of the −73 position of gypsy-like retrotransposon (AT4G07700) as control. Lower panel: Schematic illustration of positions and dynamics of nucleosomes on the AP3 promoter in wild-type, swp73a-1, and swp73b-1 plants. Gray circles represent positioned nucleosomes, and dashed circles correspond to nucleosomes that are not positioned.
(F) In situ hybridization of antisense AGL24, AP1, and AP3 transcripts in shoot apical meristems of 14-d-old wild-type, swp73a-1, and swp73b-1 plants. Note irregular shape of swp73b-1 meristem.