Fig. 2.
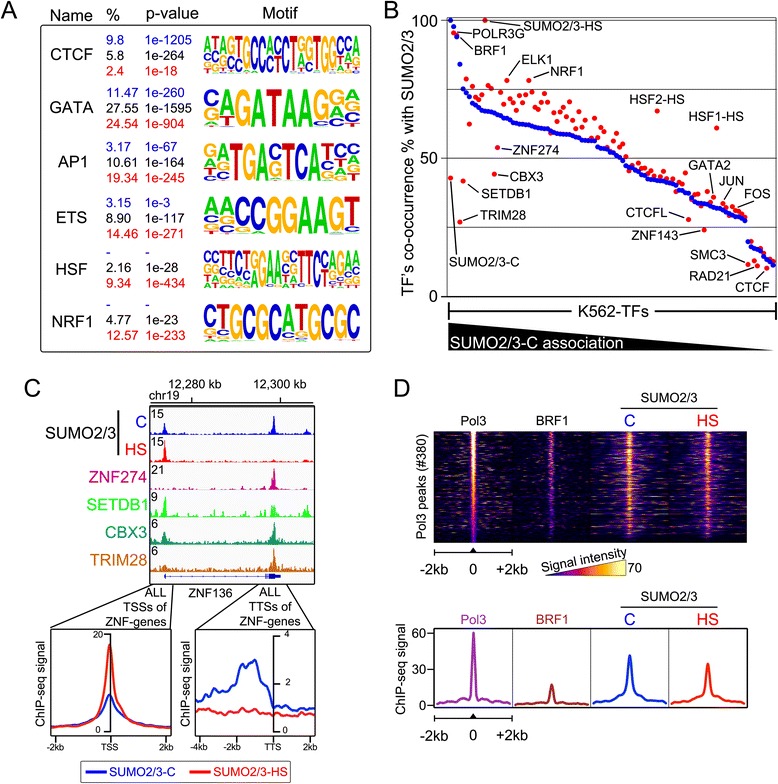
Characterization of SUMO2/3 chromatin binding sites in K562 cells. a Enrichment of DNA-binding motifs within SUMO2/3 peaks. Percentage of SUMO2/3 peaks with the motif and P value of enrichment are indicated with color code (C unique, blue; C/HS-shared, black; SUMO2/3 HS, red). b Co-occurrence of K562 TFs and SUMO2/3 peaks. Scatter plot of TF co-occurrence with SUMO2/3 C (blue dots) and SUMO2/3 HS (red dots) peaks. Y-axis represents co-occurrence percent of analyzed TF and on X-axis all analyzed TFs are arranged according to their co-occurrence percent with SUMO2/3 C. Complete list of analyzed K562-TFs and co-occurrence percentages in Table S1. c SUMO2/3 occupancy at ZNF genes. SUMO2/3 ChIP-seq track in control (C) and HS conditions aligned with ChIP-seq tracks of ZNF274, SETDB1, CMX3, and TRIM28 at ZNF136 locus. Comparison of SUMO2/3 signal at TSSs and TTSs of all annotated ZNF genes in C and HS conditions. d Association of SUMO2/3 and Pol3. Heat map of ±2 kb window centered at Pol3 binding sites and showing ChIP-seq signals of Pol3 (POLR3G subunit), Pol3-associated TF BRF1, and SUMO2/3 in C and HS conditions using false-color scale. Line profile of average ChIP-seq signals in ±2 kb window from the centers of Pol3-binding sites
