Abstract
Background
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) has been suggested to be related with the pathogenesis and progression of osteoarticular degenerations. This study therefore aimed to investigate the relationship between the polymorphism of the TRAIL gene and the pathogenesis and severity of intervertebral disc degeneration (IDD) via detection of serum TRAIL expression levels.
Material/Methods
A total of 100 IDD patients in our hospital were recruited in the experimental group, while another cohort of 100 healthy individuals was employed as the control group. Blood samples collected from all people were quantified for TRAIL level using enzyme-linked immunosorbent assay (ELISA), in addition to allele and genotype frequency analysis via fluorescent PCR for TRAIL gene.
Results
At loci 1525 and 1529 in 3′-untranslated region (UTR) of 5th exon of TRAIL gene, 3 different genotypes were identified: experimental group had higher frequency of 1525CG/1595CC, 1525G and 1595C alleles, compared to the control group (p<0.05). Patients under Schneiderman grade IV had significantly higher allele frequency compared to those at grade II or III. Serum TRAIL level was also higher in the experimental group compared to the control group, and in grade IV patients compared to grade II or III patients (p<0.05).
Conclusions
The G/C mutation at loci 1525/1595 of TRAIL gene may induce the progression of IDD, as the down-regulation of TRAIL can aggravate the severity of the disease.
Keywords: Manipulation, Spinal; Ovotesticular Disorders of Sex Development; Receptors, TNF-Related Apoptosis-Inducing Ligand
Background
As a common osteoarticular disease, intervertebral disc degeneration (IDD) affected millions of patients but without clear illustration of its pathogenesis. So far various factors including collagen, proteoglycan, vitamin D receptor, Sox9, matrix metalloproteinase and interleukin have been suggested as candidates for the occurrence of IDD [1]. It is worth noticing that many important cytokines also participate in the process of osteoarticular degeneration [2].
Tumor necrosis factor related apoptosis inducing ligand (TRAIL) is a member of the tumor necrosis factor superfamily and plays a specific role in the induction of tumor cell apoptosis, but without any significant effects on normal tissues [3–5]. Currently there are few studies regarding the genetic regulation of TRAIL and IDD. Therefore this study aims to investigate relationship between the polymorphism of TRAIL gene and occurrence and severity of IDD, in an attempt to further elucidate the pathogenesis of IDD and to provide basis for clinical treatment.
Material and Methods
Research objects
A total of 100 IDD patients with confirmed diagnosis in our hospital between October 2013 and October 2014 were recruited as the experimental group. Another cohort of 100 healthy individuals was recruited during the physical examination. In the experimental group, there were 54 males and 46 females, with aging between 31 and 81 years old (average=59 years old). Subtyping of patients revealed 38 cases of lumbar disc herniation, 25 cases of lumbar spinal stenosis and 37 cases of lumbar spinal degenerative scoliosis. The control group had 58 males and 42 females, aging between 34 and 70 years old (average=49 years old). The experimental protocol has been pre-approved by the ethical committee of our hospital and written consents have been obtained from all patients and healthy volunteers.
All IDD patients were evaluated based on Schneiderman scoring system [6] following radiological examinations: Grade I, no signal loss of intervertebral disc; Grade II, moderate signal loss of intervertebral disc; Grade III, severe signal loss of intervertebral disc; Grade IV, complete signal loss of intervertebral disc. Results showed that there were 32, 34 and 34 patients in Grade II, III and IV, respectively. All individuals in the control group belonged to Grade I. No significant difference regarding sex and age has been identified between the disease and control group.
Samples collection
4 mL fasting blood samples were collected from elbow veins of all objects and were aliquot into 2 tubes, of which 1 contains EDTA for anti-coagulation while another 1 is used for serum extraction by 3000 rpm centrifugation of 10 min. Serum was frozen at −20°C for further use.
Gene polymorphism analysis
Total DNA in the blood was extracted using genomic DNA extraction kit from EDTA-treated blood samples. The concentration and purity of DNA was further evaluated using spectrometry and agarose gel electrophoresis.
Specific primers of TRAIL (Forward, 5′-AACAT CTTCT GTCTT TATAAT C-3′; Reverse, 5′-AAATA ACACG TACTT ACTGA AG-3′) were employed along with Taq PCR pre-mixtures and genomic DNA templates for PCR amplification. A total of 30 cycles (94°C denature for 30 s, 48°C annealing for 90 s and 72°C elongation for 45 s). PCR amplification products were revealed as having a 484-bp length by agarose gel electrophoresis.
PCR products were further digested by Rsa I or Tas I restriction enzyme. In brief, 10 μL DNA fragments were mixed with 1 μL enzymes, 2 μL buffer (10X) and 16 μL deionized water. The mixture was incubated at 65°C for 3 h. The digestion products were then separated by agarose gel electrophoresis to reveal genetic polymorphism.
Serum TRAIL assay
Serum samples separated from blood was quantified for TRAIL levels using immunosorbent assay (ELISA) kit following the manual instruction. The concentration in samples was calculated based on the standard curve.
Statistical analysis
SPSS 17.0 software package was used to analyze all collected data, of which enumeration data were compared by chi-square test while measurement data was tested by analysis of variance (ANOVA). Between-group-comparison was performed by least significant difference (LSD) method. A statistical significance was defined when p<0.05.
Results
Genotyping of TRAIL gene
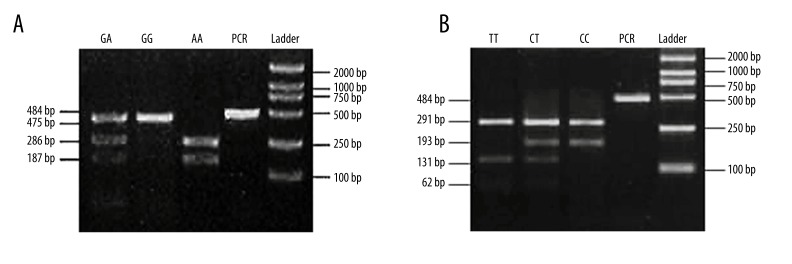
After differential restriction enzyme digestion, we found gene polymorphisms in both loci 1525 and loci 1595 of TRAIL gene. Using Rsa I digestion, 3 different genotypes, GG, AA and GA were found at loci 1525 (Figure 1A). After Tas I processing, CC, TT and CT genotypes were also revealed at loci 1595 (Figure 1B).
Figure 1.
Phenotyping of TRAIL genes. (A) and (B) showed enzymatic digestion of PCR products as revealed by agarose gel electrophoresis. (A), the digestion by Rsa I found 3 genotypes (GA, GG and AA) at loci 1525; (B), Tas I enzyme produced 3 genotypes (TT, CT and CC).
Comparisons of genotype and allele frequency of TRAIL gene between groups
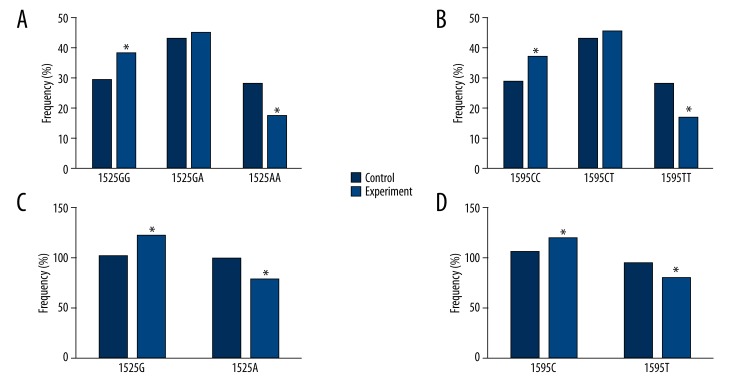
Frequencies of both genotypes and alleles at loci 1525and loci 1595 were analyzed between the experimental and the control group. As shown in Figure 2A, 2B, the frequency of 1525GG and 1595CC genotypes was significantly elevated in the experimental group (38% and 37% for loci 1525 and 1595, respectively) when compared to the control group (29% for both loci; p<0.05 as shown by chi-square test). Moreover, when we compared the allele frequency, it was found that experimental group had significantly higher frequency of 1525G (61%) and 1595C (60%) compared to those in the control group (51% and 53% for loci 1525 and 1595, respectively; p<0.05 using chi-square test) as shown in Figure 2C, 2D.
Figure 2.
Genotype and allele frequency of TRAIL gene. (A) and (B) showed genotyping analysis of TRAIL gene at loci 1525 and 1595, respectively. Experimental group had higher frequency of 1525GG and 1595CC, and lower frequency of 1525AA and 1595TT, when compared to the control group. (C) and (D) showed allele frequency of TRAIL gene at those 2 loci. Frequency of 1525G and 1595C was elevated in the experimental group. *, p<0.05 using chi-square test.
Analysis of TRAIL gene polymorphism among different grades of IDD patients
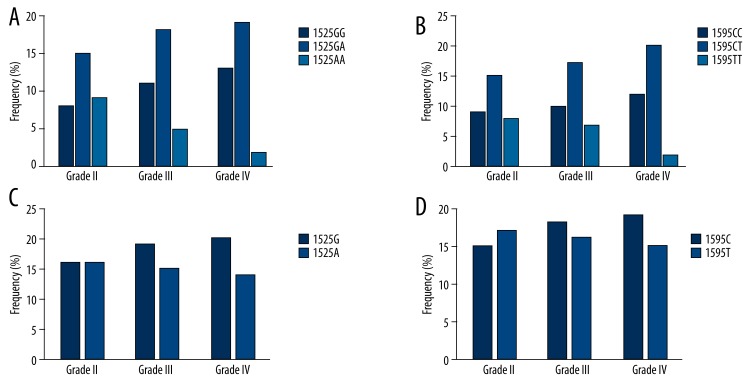
We further analyzed the gene polymorphism pattern of TRAIL gene among different clinical grades of IDD. Results (Figure 3) showed that patients with higher grade IDD had more genotypes including 1525GG (Figure 3A) and 1595CC (Figure 3B), along with alleles 1525G (Figure 3C) and 1595C (Figure 3D), although such differences were of insignificant differences under chi-square test.
Figure 3.
TRAIL gene polymorphism among IDD grades. (A) and (B) showed genotype frequency at loci 1525 and 1595, respectively. Higher grade patients had more 1525GG and 1595CC. (C) and (D) plotted allele frequency at those 2 loci. More 1525G and 1595C alleles occurred in higher grade IDD patients.
TRAIL gene polymorphism and serum TRAIL levels
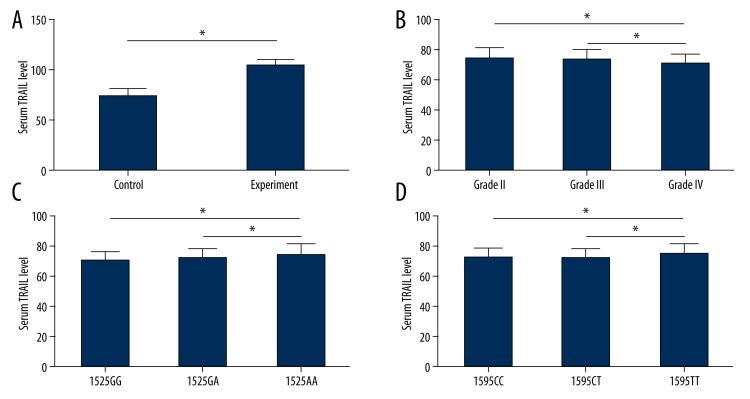
The serum level of TRAIL was also quantified in both experimental and control individuals. Results showed a significantly elevated serum TRAIL expression level in the experimental group when compared to the control group (103.7±6.1 vs. 74.1±7.0; Figure 4A; p<0.05 using t-test). A further analysis was made among different clinical grades of IDD and revealed that Grade IV patients had significantly lower TRAIL levels (70.1±6.4) compared to Grade II (74.1±7.0) or Grade III (72.7±6.8) ones (Figure 4B; p<0.05 using post-hoc LSD test).
Figure 4.
Serum TRAIL levels and gene polymorphism. (A) and (B) showed serum TRAIL levels between control and experimental groups (A), or among different grade of IDD patients (B). Disease individuals had higher serum TRAIL levels, while patients with more advanced grade had lower protein expressions. (C) and (D) showed TRAIL protein levels among different genotypes at loci 1525 and 1595, respectively. Patients with 1525AA or 1595TT of TRAIL gene tends to have higher serum TRAIL levels. *, p<0.05 using post-hoc least significant difference (LSD) test.
In an attempt to find any relationship between TRAIL gene polymorphism and serum protein level, we found that IDD patients with 1525AA genotype had higher serum TRAIL level (74.5±7.0) compared to 1525GG (70.1±6.3) or 1525GA (72.1±6.5) ones (Figure 3C; p<0.05 using post-hoc LSD test). At loci 1595, similar pattern also occurred, as 1595TT individuals had higher serum TRAIL level (75.4±6.2) when compared to 1595CC (72.1±6.3) or 1595CT (72.3±6.5) ones (Figure 4D; p<0.05 using post-hoc LSD test).
Discussion
Lumbar intervertebral disc degeneration is manifested with chronic pains in waist and legs, numbness and fatigue [7,8]. Previous studies suggested the participation of cytokines as critical inflammatory factors in the occurrence and progression of degenerative osteoarticular disease and osteoarthritis [9]. For example, cytokines can induce phagocytes in the fibrous ring and nucleus pulposus cells of the intervertebral disc [10,11].
TRAIL works as an important immune regulatory factor as it is involved in the body’s immune modulation, immune homeostasis and immune surveillance [12,13]. Multiple tissues and organs are sensitive to TRAIL-induced cell apoptosis [14,15]. The polymorphism of TRAIL gene was firstly proposed by Strekalova et al., who suggested that such polymorphism is not related with the protein expression level, and the mutation at loci 1525 and 1595 may play a role in the pathogenesis and progression of lumbar IDD [16]. This study therefore further explored the relationship between TRAIL gene polymorphism at loci 1525/1595 and the pathogenesis of IDD, in order to provide basis for clinical treatment.
Current knowledge about TRAIL-related disease mainly involves tumor, auto-immune disease, and viral infections. Studies have shown predisposed gene loci of fat liver and systematic lupus erythematosus (SLE) at 1525G/A and 1595C/T of 3′-UTR of 5th exon of TRAIL gene. There were also reports about the genetic polymorphism of TRAIL gene and ovary cancer, as well as the role of TRAIL in stomach cancer cell apoptosis. Past studies indicated significantly suppressed TRAIL protein expression level in malignant ovary epithelial tumor cells, when compared to those in benign ovary tumor and normal tissues. Further analysis identified lower TRAIL protein in stage III/IV ovary cancer compared to those in stage I/II. The application of 1100 mg/L recombinant human soluble TRAIL protein can induce the apoptosis of human ovary cancer cell 3AO, suggesting the relationship between TRAIL and oncogenesis/progression of ovary cancer. The multiple sclerosis (MS) has also been suggested to be related with the disease allele at 5th exon of TRAIL gene at loci 1595. Kikuchia et al. studied the correlation between TRAIL and pathogenesis of MS and found that the C allele at loci 1595 is a risk factor of MS. TRAIL may play different roles in various diseases, as it can exert unique function via the induction of cell apoptosis or arresting cell cycle. Elevated TRAIL mRNA levels have been found in peripheral blood-derived monocytes in SLE patients, in addition to elevated serum TRAIL level. These phenotypes were accompanied with facilitated release of functional TRAIL from T cells, as stimulated by IFN-α. The TRAIL-induced cell apoptosis may further potentiate the programmed death of abnormal cells in SLE patients. In other studies about Graves’s disease, TRAIL expression in cultured lymphocytes were elevated with higher concentration of thyroid hormones, inducing higher apoptotic rates of cells, making the follicular cells of thyroid to escape from T cell-inducted cytotoxicity. Therefore, lower apoptosis of follicular cells occurred in Graves’s disease, along with increased infiltrated lymphocytes, further aggravating proliferation and hyperfunction of thyroid.
In this study, a genetic analysis revealed the existence of susceptible gene at 3′-UTR region of TRAIL gene. A total of 3 genotypes existed at both loci: 1525GG, 1525GA, 1525AA, 1595CC, 1595CT, and 1595TT (Figure 1). IDD patients had higher 1525GG/1595CC and 1525GA/1595CT phenotype frequency, and higher 1525G/1595C allele frequency when compared to healthy individuals (Figure 2). This is consistent with a previous study suggesting an elevated rate of 1525G/1595C alleles in disease tissues compared to control ones [17,18]. All these results suggest that the high expression of 1525G/1595C in 3′-UTR of 5th exon of TRAIL gene may be related to the pathogenesis of lumbar IDD. The detailed mechanisms, however, remain to be fully elucidated.
This study, for the first time, discovered the differential distribution of genotype/allele frequencies of TRAIL gene among different clinical grades of IDD patients. Grade IV patients had higher frequency of 1525GG/1595CC and 1525G/1595C compared to Grade II or Grade III individuals (Figure 3). The occurrence rate of 1525AA/1595TT and 1525A/1595T was depressed in advanced-stage patients. These results point the possible value of TRAIL gene polymorphism in predicting the risk and severity of IDD, although its underlying mechanism remains unknown. Related studies of TRAIL in ulcerative colitis defined the loci 1525 and 1595 in a single haplotype, as these 2 loci showed complete linkage [19, 20]. This study confirmed this opinion by showing complete linkage of genotypes at loci 1525 and 1595.
It is interesting that lower serum TRAIL protein levels were observed in the experimental group and more advanced IDD patients had much lower protein levels (Figure 4A, 4B). These results suggest that lower TRAIL expression may be related with lumbar IDD. Therefore, serum TRAIL may work as an index for predicting the severity and prognosis of IDD. A further study revealed that individuals with 1525GG/1595CC genotypes had lower serum TRAIL levels compared to those with 1525AA/1525TT genotypes (Figure 4C, 4D). This is in an agreement with our finding that 1525GG/1595CC individuals are predisposed to more advanced IDD. Therefore, a substantial relationship exists among TRAIL genotype, protein level, and disease severity, making TRAIL a potential candidate for both diagnosis and treatment of IDD.
This study has its inherent weakness and limitations. The detailed mechanism requires further studies for confirmation of findings. No correlation studies have been performed between the different genotypes of TRAIL and clinical manifestations. The sample number of this study is also limited; thus, requiring larger follow-up studies that include samples from different regions and ethnic groups.
Conclusions
In summary, a correlation exists between the polymorphism of TRAIL gene and lumbar IDD. The 1525G/1595C alleles at 3′-UTR of 5th exon of TRAIL gene may induce the pathogenesis and progression of lumbar IDD. Moreover, those alleles can lead to the suppressed expression of TRAIL protein, further aggravating the condition of IDD. Our results support the potential value of TRAIL gene polymorphism and serum levels in predicting disease stage and prognosis, although more evidence from studies with larger sample sizes and animal studies are required to further elucidate its detailed mechanisms.
Footnotes
Source of support: Departmental sources
References
- 1.Erkul E, Kucukodaci Z, Pinar D, et al. Trail and trail receptors in patients with laryngeal cancer. Head Neck. 2015 doi: 10.1002/hed.24035. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 2.Bisgin A, Yalcin AD, Gorczynski RM. Circulating soluble tumor necrosis factor related apoptosis inducing-ligand (TRAIL) is decreased in type-2 newly diagnosed, non-drug using diabetic patients. Diabetes Res Clin Pract. 2012;96(3):e84–86. doi: 10.1016/j.diabres.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 3.He X, Chen X, Zhang X, et al. An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res. 2015;43(7):3712–25. doi: 10.1093/nar/gkv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurne A, Guc D, Canpinar H, et al. Analysis of BAFF and TRAIL expression levels in multiple sclerosis patients: evaluation of expression under immunomodulatory therapy. Acta Neurol Scand. 2011;123(1):8–12. doi: 10.1111/j.1600-0404.2010.01346.x. [DOI] [PubMed] [Google Scholar]
- 5.Wiersma VR, de Bruyn M, Shi C, et al. C-type lectin-like molecule-1 (CLL1)-targeted TRAIL augments the tumoricidal activity of granulocytes and potentiates therapeutic antibody-dependent cell-mediated cytotoxicity. MAbs. 2015;7(2):321–30. doi: 10.1080/19420862.2015.1007811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu N, Chen T2, Wang X, et al. Msi1 confers resistance to TRAIL by activating ERK in liver cancer cells. FEBS Lett. 2015;589(8):897–903. doi: 10.1016/j.febslet.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 7.Dao P, Smith N, Scott-Algara D, et al. Restoration of TRAIL-induced apoptosis in resistant human pancreatic cancer cells by a novel FAK inhibitor, PH11. Cancer Lett. 2015;360(1):48–59. doi: 10.1016/j.canlet.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 8.Kisim A, Atmaca H, Cakar B, et al. Pretreatment with AT-101 enhances tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis of breast cancer cells by inducing death receptors 4 and 5 protein levels. J Cancer Res Clin Oncol. 2012;138(7):1155–63. doi: 10.1007/s00432-012-1187-1. [DOI] [PubMed] [Google Scholar]
- 9.Yerbes R, Lopez-Rivas A. Itch/AIP4-independent proteasomal degradation of cFLIP induced by the histone deacetylase inhibitor SAHA sensitizes breast tumour cells to TRAIL. Invest New Drugs. 2012;30(2):541–47. doi: 10.1007/s10637-010-9597-x. [DOI] [PubMed] [Google Scholar]
- 10.Guzmán EA, Johnson JD, Linley PA, et al. A novel activity from an old compound: Manzamine A reduces the metastatic potential of AsPC-1 pancreatic cancer cells and sensitizes them to TRAIL-induced apoptosis. Invest New Drugs. 2011;29(5):777–85. doi: 10.1007/s10637-010-9422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HB, Kim MJ, Lee SH, et al. Amurensin G, a novel SIRT1 inhibitor, sensitizes TRAIL-resistant human leukemic K562 cells to TRAIL-induced apoptosis. Biochem Pharmacol. 2012;84(3):402–10. doi: 10.1016/j.bcp.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Zhu X, Zhang K, Wang Q, et al. Cisplatin-mediated c-myc overexpression and cytochrome c (cyt c) release result in the up-regulation of the death receptors DR4 and DR5 and the activation of caspase 3 and caspase 9, likely responsible for the TRAIL-sensitizing effect of cisplatin. Med Oncol. 2015;32(4):1–15. doi: 10.1007/s12032-015-0588-9. [DOI] [PubMed] [Google Scholar]
- 13.Kauntz H, Bousserouel S, Gossé F, Raul F. The flavonolignan silibinin potentiates TRAIL-induced apoptosis in human colon adenocarcinoma and in derived TRAIL-resistant metastatic cells. Apoptosis. 2012;17(8):797–809. doi: 10.1007/s10495-012-0731-4. [DOI] [PubMed] [Google Scholar]
- 14.Gonelli A, Radillo O, Drioli S, et al. Pegylated TRAIL retains anti-leukemic cytotoxicity and exhibits improved signal transduction activity with respect to TRAIL. Invest New Drugs. 2012;30(2):828–32. doi: 10.1007/s10637-010-9599-8. [DOI] [PubMed] [Google Scholar]
- 15.Xu L, Zhang Y, Liu J, et al. TRAIL-activated EGFR by Cbl-b-regulated EGFR redistribution in lipid rafts antagonises TRAIL-induced apoptosis in gastric cancer cells. Eur J Cancer. 2012;48(17):3288–99. doi: 10.1016/j.ejca.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Strekalova E, Malin D, Good DM, Cryns VL. Methionine deprivation induces a targetable vulnerability in triple-negative breast cancer cells by enhancing TRAIL receptor-2 expression. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-14-2792. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazurek N, Byrd JC, Sun Y, et al. Cell-surface galectin-3 confers resistance to TRAIL by impeding trafficking of death receptors in metastatic colon adenocarcinoma cells. Cell Death Differ. 2012;19(3):523–33. doi: 10.1038/cdd.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szliszka E, Czuba ZP, Bronikowska J, et al. Ethanolic extract of propolis augments TRAIL-induced apoptotic death in prostate cancer cells. Evid Based Complement Alternat Med. 2011;2011:535172. doi: 10.1093/ecam/nep180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hori T1, Kondo T, Kanamori M, et al. Ionizing radiation enhances tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis through up-regulations of death receptor 4 (DR4) and death receptor 5 (DR5) in human osteosarcoma cells. J Orthop Res. 2010;28(6):739–45. doi: 10.1002/jor.21056. [DOI] [PubMed] [Google Scholar]
- 20.Woo JK, Kang JH, Jang YS, et al. Evaluation of preventive and therapeutic activity of novel non-steroidal anti-inflammatory drug, CG100649, in colon cancer: Increased expression of TNF-related apoptosis-inducing ligand receptors enhance the apoptotic response to combination treatment with TRAIL. Oncol Rep. 2015;33(4):1947–55. doi: 10.3892/or.2015.3793. [DOI] [PubMed] [Google Scholar]