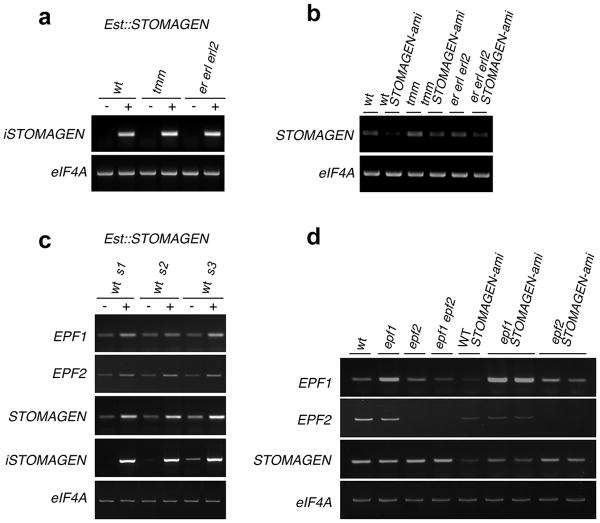
Extended Data Fig. 2. RT-PCR analysis of STOMAGEN transcripts in transgenic lines used in this study.
(a) Expression of estradiol-inducible STOMAGEN transgene (iSTOMAGEN) in transgenic lines expressing estradiol-inducible STOMAGEN overexpression (Est∷STOMAGEN) lines from wild-type (wt), tmm, and er erl1 erl2 triple mutant background with or without estradiol induction. (b) Expression of the endogenous STOMAGEN transcripts in each genotype carrying STOMAGEN-ami construct. tmm or er erl1 erl2 mutation does not seem to affect STOMAGEN transcript levels. (c) Expression of EPF1, EPF2, total STOMAGEN, and STOMAGEN transgene (iSTOMAGEN) transcripts in transgenic Est∷STOMAGEN lines (in 2 different T1 populations [s1 and s2] and a representative T3 line [s3]) with or without estradiol induction. iSTOMAGEN-OX causes modest increase in EPF1 and EPF2 transcripts, which accords with increased stomatal differentiation by iSTOMAGEN. (d) EPF1, EPF2, and STOMAGEN transcript accumulation in wild-type (wt) and single- and higher-order loss-of-function mutants of epf1, epf2, and stomagen (STOMAGEN-ami). For epf1 STOMAGEN-ami and epf2 STOMAGEN-ami lines, two different F3 populations derived from the same genetic crosses were used to test the reproducibility. STOMAGEN expression is not influenced by epf1 and epf2 mutations, consistent with the proto-mesophyll expression of STOMAGEN. On the other hand, EPF2 expression is reduced by STOMAGEN-ami, consistent with reduced stomatal cell lineages by STOMAGEN cosuppression. As reported, epf1 has a T-DNA insertion within the 5′UTR12, which results in accumulation of aberrant transcripts. For all experiments, elF4A was used as a control. For primer sequences see Extended Data Table 1.