Summary
Background
We evaluated 10 single-nucleotide polymorphisms (SNPs) identified in three European case–control studies as risk factors for venous thrombosis.
Objectives
We sought to replicate the positive findings from this report among Whites and to evaluate the association of these SNPs with venous thrombosis for the first time among Blacks.
Patient/methods
These SNPs were evaluated in a case–control study of deep vein thrombosis and pulmonary embolism that included 1076 cases and 1239 controls. About 50% of subjects were African Americans. We measured plasma factor (F) XI on a subset of subjects.
Results
Among Whites, positive findings for rs13146272 in the CYP4V2 gene, for rs3087505 in the KLKB1 gene and for rs3756008 and rs2036914 in the F11 gene were found. We did not find significant associations for rs2227589 in the SERPINC1 gene and for rs1613662 in the GP6 gene. Among Blacks, rs2036914 in F11 and rs670659 in RGS7 were related to venous thrombosis, but the study had limited statistical power for many SNPs. Among Blacks, plasma FXI was related to two SNPs and the OR relating to the 90th percentile of the control distribution of plasma FXI was 2.6 (95% CI, 1.4, 5.0).
Conclusions
Our study supports the finding that genetic variants in the F11 gene are risk factors for venous thrombosis among both Whites and Blacks, although the findings in Blacks require confirmation. A meta-analysis of five case–control studies indicates that rs2227589 in the SERPINC1 gene, rs13146272 in the CYP4V2 gene and rs1613662 in the GP6 gene are risk factors for venous thrombosis among Whites.
Keywords: Blacks, genetics, pulmonary embolism, thrombosis
Introduction
While several recent studies have identified single-nucleotide polymorphisms (SNPs) linked to venous thrombosis embolism (VTE) in predominantly White populations, identification of genetic variants associated with VTE in Blacks has had little success. This is so largely because of the paucity of Black subjects in epidemiologic studies of VTE. In a recent study from the Netherlands, Bezemer et al. [1] evaluated 19 682 SNPs on 10 887 genes in three case–control studies of first deep-vein thrombosis (DVT). Using a sequential strategy, they selected nine SNPs that were assayed in a third case–control study. From this case–control study they identified three SNPs that were associated with DVT (rs13146272 in the CYP4V2 gene, rs2227589 in the SERPINC1 gene and rs1613662 in the GP6 gene).
The purpose of this report is to evaluate these three SNPs as well as four other variants in the vicinity of rs13146272 (rs3756008 in F11; rs3087505 in KLKB1; rs2036914 in F11; and rs4253418 in F11) in a case–control study of VTE that included both Whites and Blacks. We also evaluated three other SNPs identified by Bezemer et al. with false discovery rates < 0.20 (rs670659 in RGS7, rs2001490 in NR1I2 and rs1523127 in NAT8B) and measured plasma factor (F) XI in a subset of subjects.
Materials and methods
The Genetic Attributes and Thrombosis Epidemiology (GATE) study methods have been described elsewhere [2]. Briefly, VTE cases are White or Black persons between the ages of 18 and 82 with a recently diagnosed episode of a deep vein thrombosis (DVT) or pulmonary embolism (PE) hospitalized at one of two university hospitals in Atlanta, Georgia, USA. VTE includes idiopathic cases for which there was no identified provocation for their VTE (those arising in the absence of cancer, surgery, a recent traumatic injury or prolonged immobilization; those arising during pregnancy; or those arising in conjunction with the placement of a central venous catheter) and cases with any of these risk factors. All diagnoses were confirmed by objective imaging studies.
Control subjects were sampled from a list of patients who had visited the office of one of ten physicians at two university-affiliated primary care practises. Persons with a history of VTE or those currently taking anticoagulant medications were not eligible as controls. An attempt was made to select controls such that they would be similar to cases with respect to sex, ethnicity and age.
Cases were interviewed in the hospital and controls at the CDC. A whole blood sample was obtained from all subjects. TaqMan allelic-discrimination assay designed around a single-nucleotide polymorphism (SNP) was used to genotype SNPs (Applied BioSystems, Foster City, CA, USA). The CYP4V2 Lys259Gln G/A substitution (rs13146272), SERPINC1 G/A (rs2227589; NM_000488.2) intronic substitution, GP6 Ser219-Pro A/G substitution (rs16113662; NM_001083899.1), FXI A/T (rs3756008), KLKB1 (rs3087505), FXI C/T (rs2036914), FXI A/G (rs4253418) SNPs and the RGS7 C/T (rs670659 – NM_002924.4) intronic substitution were determined by Taqman Genotyping Assays. The NR1I2 C/A (rs1523127; NM_033013.2) was determined by the TaqMan Drug Metabolism Genotyping Assay. For NAT8B Gly112Ala (rs2001490; GGT-GCT substitution) a custom-made TaqMan SNP Genotyping Assay AH1RDQA was used. FIX Thr194Ala (ACT-GCT; rs6048; NM_000133.3) also was determined by TaqMan Genotyping. All individual Taqman Genotyping assay results were validated with 5–10% sequencing results. We used the same nomenclature and labeling of ‘risk alleles’ for each SNP as that defined in Bezemer et al., where the risk allele may be the minor or major allele for that SNP.
The Total Human Coagulation Factor XI Antigen Assay Kit fromMolecular Innovations (Novi, MI, USA) was used to determine the total FXI antigen (FXI and FXIa) concentration in the plasma samples. The assay was performed as directed by the manufacturer and plasma samples were diluted 1000× in the blocking buffer (0.1 M Tris, 0.15 M NaCl, pH 7.4 with 3% BSA). The A450 nm was measured with a MAXline Microplate Reader fromMolecular Devices (Sunnyvalle, CA, USA).
We randomly sampled 500 subjects to measure plasma FXI. For the White controls we selected subjects such that each of the three genotypes for rs2036914 would have about 50 subjects. For Black cases we selected 100 with idiopathic disease and available plasma and for Black controls we selected 250. With these sample sizes we estimated that we had more than 80% power to detect an association between FXI and the genotypes of rs2036914 and rs3756008 among White controls based upon the magnitude of the associations reported by Bezemer et al. [1]. Our power was> 80% to detect a difference in plasma FXI between Black cases and controls assuming that their difference would be about the same as that in the Dutch study [3]. We obtained plasma FXI measurements on 498 subjects (152 White controls, 248 Black controls and 98 Black cases).
Hardy–Weinberg equilibrium was evaluated in controls through the use of likelihood ratio tests [4]. We used the correlation coefficient as a measure of linkage disequilibrium among the SNPs [5]. We obtained the odds of genotypes with one or two risk alleles compared with the genotype homozygous for the absence of the risk allele among cases and divided by the corresponding odds among controls. This odds ratio (OR) estimates the relative risk of VTE. To evaluate trend in risk, we assumed an additive increase in log odds in a logistic regression model for each additional risk allele. Odds ratios, 95% confidence limits and two-tailed P values were obtained by logistic regression [6]. To control the Type I error rate for multiple comparisons, we used the false discovery rate (FDR) and its associated measure, the q value [7,8]. The FDR is a function of the P value and the number of tests performed. The FDR is the proportion of tests judged significant that are truly null (false positives). Thus, if the FDR is set at 0.10, all tests with a q less than 0.10 will be judged significant and we expected that 10% of them would be false positives. A q value of say, 0.08, means that if tests with a q < 0.08 are judged as significant, we would expect that about 8% of them would be false positives.
We used the logarithm of the plasma FXI measurements in our statistical analyses because the unconverted measurements were skewed right. We used linear regression on the logarithmic scale to evaluate the trend of themeasurements across the three genotypes for each SNP and to evaluate the mean difference between Black cases and controls.
We performed a meta-analysis on three SNPs evaluated in five studies (including the present study). We assumed a fixed effects model and computed odds ratios based on alleles (odds of risk allele among cases divided by the odds of the risk allele among controls). We used logistic regression to estimate the overall summary allelic odds ratios with 95% CIs and to evaluate whether they were homogenous across studies. All analyses were implemented through the use of the Stata software [9].
This study was approved by the Institutional Review Boards of Emory University, Atlanta, GA, USA, and the Centers for Disease Control and Prevention.
Results
The study included 1076 cases and 1239 controls. Five hundred and thirty (49%) cases and 575 (46%) controls were Blacks. Five hundred and forty (50%) cases and 611 (49%) controls were men. The median age and interquartile range for cases was 50 (39, 59) while that for controls was 51 (40, 60). Cases included 641 provoked and 435 idiopathic VTEs. Of the 641 provoked VTEs, 282 had cancer.
The distribution of the genotypes for the three statistically significant SNPs in the Bezemer et al. report, for the three SNPs in the vicinity of rs13146272 (SNP rs4253418 could not be analyzed because all but one subject was homozygous for the major allele) and for the three SNPs of marginal statistical significance is displayed in Table 1. The genotypes associated with each of these SNPs were in Hardy–Weinberg equilibrium among both White and Black controls.
Table 1.
Distribution of genotypes among cases and controls with odds ratios, 95% confidence limits and the trend odds ratio with adjusted and unadjusted P and q values for Whites and Blacks
| Gene/SNP/chromosome | Cases | Controls | OR | 95% CI |
|---|---|---|---|---|
| Whites | ||||
| CYP4V2/rs13146272/4 | ||||
| CC | 67 | 102 | 1.0 | – |
| CA | 233 | 303 | 1.2 | 0.82, 1.7 |
| AA | 244 | 256 | 1.5 | 1.0, 2.1 |
| Prev(A) | 0.66 | 0.62 | Trend | |
| OR = 1.21; | ||||
| P = 0.02; | ||||
| q* = 0.08 | ||||
| KLKB1/rs3087505/4 | ||||
| TT | 1 | 5 | 1.0 | – |
| TC | 81 | 126 | 3.2 | 0.37, 28.0 |
| CC | 463 | 533 | 4.3 | 0.51, 37.3 |
| Prev(C) | 0.92 | 0.90 | Trend | |
| OR = 1.40; | ||||
| P = 0.02 | ||||
| q = 0.08 | ||||
| F11/rs2036914/4 | ||||
| TT | 100 | 156 | 1.0 | – |
| TC | 267 | 335 | 1.2 | 0.92, 1.7 |
| CC | 177 | 170 | 1.6 | 1.2, 2.3 |
| Prev(C) | 0.57 | 0.51 | Trend | |
| OR = 1.28; | ||||
| P = 0.003 | ||||
| q = 0.03 | ||||
| F11/rs3756008/4 | ||||
| AA | 165 | 265 | 1.0 | – |
| AT | 265 | 298 | 1.4 | 1.1, 1.8 |
| TT | 115 | 101 | 1.8 | 1.3, 2.5 |
| Prev(T) | 0.45 | 0.38 | Trend | |
| OR = 1.36; | ||||
| P < 0.001 | ||||
| q = 0.003 | ||||
| SERPINC1/rs2227589/1 | ||||
| CC | 426 | 530 | 1.0 | – |
| CT | 113 | 127 | 1.1 | 0.83, 1.5 |
| TT | 5 | 7 | 0.89 | 0.28, 2.8 |
| Prev(T) | 0.11 | 0.11 | Trend | |
| OR = 1.07; | ||||
| P > 0.20 | ||||
| GP6/rs1613662/19 | ||||
| GG | 16 | 17 | 1.0 | – |
| GA | 134 | 175 | 0.81 | 0.40, 1.7 |
| AA | 396 | 471 | 0.89 | 0.45, 1.8 |
| Prev(A) | 0.85 | 0.84 | Trend | |
| OR = 1.04; | ||||
| P > 0.20 | ||||
| RGS7/rs670659/1 | ||||
| TT | 75 | 88 | 1.0 | – |
| TC | 240 | 314 | 0.90 | 0.63, 1.3 |
| CC | 225 | 256 | 1.0 | 0.72, 1.5 |
| Prev(C) | 0.64 | 0.63 | Trend | |
| OR = 1.05; | ||||
| P > 0.20 | ||||
| NR1I2/RS1523127/2 | ||||
| AA | 197 | 229 | 1.0 | – |
| AC | 253 | 325 | 0.90 | 0.70, 1.2 |
| CC | 92 | 110 | 0.97 | 0.69, 1.4 |
| Prev(C) | 0.40 | 0.41 | Trend | |
| OR = 0.97; | ||||
| P > 0.20 | ||||
| NAT8B/rs2001490/3 | ||||
| GG | 205 | 272 | 1.0 | – |
| GC | 258 | 294 | 1.2 | 0.91, 1.5 |
| CC | 77 | 91 | 1.1 | 0.79, 1.6 |
| Prev(C) | 0.38 | 0.36 | Trend | |
| OR = 1.08; | ||||
| P > 0.20 | ||||
| Blacks | ||||
| CYP4V2/rs13146272/4 | ||||
| CC | 70 | 84 | 1.0 | – |
| CA | 264 | 278 | 1.1 | 0.80, 1.6 |
| AA | 195 | 210 | 1.1 | 0.77, 1.6 |
| Prev(A) | 0.62 | 0.61 | Trend | |
| OR = 1.04; | ||||
| P > 0.20 | ||||
| KLKB1/rs3087505/4 | ||||
| TT | 2 | 3 | 1.0 | – |
| TC | 80 | 82 | 1.5 | 0.24, 9.0 |
| CC | 448 | 489 | 1.4 | 0.23, 8.3 |
| Prev(C) | 0.92 | 0.92 | Trend | |
| OR = 0.96; | ||||
| P > 0.20 | ||||
| F11/rs2036914/4 | ||||
| TT | 69 | 91 | 1.0 | – |
| TC | 217 | 257 | 1.1 | 0.78, 1.6 |
| CC | 243 | 224 | 1.4 | 1.0, 2.1 |
| Prev(C) | 0.66 | 0.62 | Trend | |
| OR = 1.22; | ||||
| P = 0.02; | ||||
| q = 0.08 | ||||
| F11/rs3756008/4 | ||||
| AA | 259 | 301 | 1.0 | – |
| AT | 227 | 234 | 1.1 | 0.88, 1.4 |
| TT | 43 | 38 | 1.3 | 0.82, 2.1 |
| Prev(T) | 0.30 | 0.27 | Trend | |
| OR = 1.13; | ||||
| P = 0.18; | ||||
| q > 0.20 | ||||
| SERPINC1/rs2227589/1 | ||||
| CC | 469 | 499 | 1.0 | – |
| CT | 58 | 71 | 0.87 | 0.60, 1.3 |
| TT | 2 | 5 | 0.43 | 0.08, 2.2 |
| Prev(T) | 0.06 | 0.07 | Trend | |
| OR = 0.83; | ||||
| P > 0.20 | ||||
| GP6/rs1613662/19 | ||||
| GG | 27 | 39 | 1.0 | – |
| GA | 197 | 198 | 1.4 | 0.85, 1.2 |
| AA | 305 | 337 | 1.3 | 0.78, 2.2 |
| Prev(A) | 0.76 | 0.76 | Trend | |
| OR = 1.02; | ||||
| P > 0.20 | ||||
| RGS7/rs670659/1 | ||||
| TT | 20 | 29 | 1.0 | – |
| TC | 160 | 203 | 1.1 | 0.62, 2.1 |
| CC | 343 | 337 | 1.5 | 0.82, 2.7 |
| Prev(C) | 0.81 | 0.77 | Trend | |
| OR = 1.26; | ||||
| P = 0.03; | ||||
| q = 0.09 | ||||
| NR1I2/RS1523127/2 | ||||
| AA | 24 | 33 | 1.0 | – |
| AC | 174 | 178 | 1.3 | 0.76, 2.4 |
| CC | 326 | 360 | 1.2 | 0.72, 2.2 |
| Prev(C) | 0.79 | 0.79 | Trend | |
| OR = 1.01; | ||||
| P > 0.20 | ||||
| NAT8B/rs2001490/3 | ||||
| GG | 83 | 109 | 1.0 | – |
| GC | 250 | 270 | 1.2 | 0.87, 1.7 |
| CC | 184 | 180 | 1.3 | 0.94, 1.9 |
| Prev(C) | 0.60 | 0.56 | Trend | |
| OR = 1.15; | ||||
| P = 0.11; | ||||
| q > 0.20 | ||||
The q value for the FDR (see text). If the P value is > 0.20 so is the q value and therefore this is not listed. SNP, single-nucleotide polymorphism.
Among Whites, the rs13146272 SNP in the CYP4V2 gene was statistically significantly (P = 0.02) associated with VTE, with a trend OR for the three genotypes of 1.21 (Table 1), similar to the OR of 1.24 reported by Bezemer et al. After adjustment for multiple comparisons, the q value for this SNP was 0.08. The three SNPs in the region of rs13146272 in the KLKB1 and F11 genes (rs3087505, rs2036914 and rs3756008) also were statistically significantly associated with VTE. After adjustment for multiple comparisons, the q values for these three SNPs were 0.08, 0.03 and 0.003, respectively. We did not find any statistically significant associations between the SNPs in the SERPINC1 and GP6 genes as had Bezemer et al. The four SNPs on chromosome 4 are in linkage disequilibrium (Table 2). Inclusion of these four SNPs in a single model eliminated the statistical significance of all except for rs3756008 in F11 (P = 0.03).
Table 2.
The correlation coefficient as a measure of linkage disequilibrium for four single-nucleotide polymorphisms (SNPs) on chromosome 4 among controls for Whites and Blacks
| SNP | rs13146272 | rs3087505 | rs2036914 | rs3756008 |
|---|---|---|---|---|
| rs13146272 | – | 0.39* | 0.33 | 0.29 |
| rs3087505 | 0.30† | – | 0.33 | 0.24 |
| rs2036914 | 0.05‡ | 0.30 | – | 0.63 |
| rs3756008 | 0.29 | 0.17 | 0.20 | – |
Entries above the main diagonal pertain to White controls.
Entries below the main diagonal pertain to Black controls.
All measures of linkage disequilibrium are statistically significant (P < 0.0001) with this exception.
Among Blacks, rs2036914 in F11 and rs670659 in RGS7 were statistically significantly associated with VTE (Table 1) before adjustment for multiple comparisons. After adjustment, the q values were 0.08 and 0.09, respectively. As for Whites, the four SNPs on chromosome 4 were in linkage disequilibrium expect for rs13146272 and rs2036914 (Table 2). The rs2036914 SNP in F11 was statistically significant in a multivariate model that included the four SNPs on chromosome 4 (P = 0.02).
The prevalence of the risk alleles among White and Black controls was statistically significantly different (P < 0.05), except for rs13146272 in CYP4V2. The q value based on the FDR method also was less than 5% for all SNPs other than rs13146272 in CYP4V2.
Among the 152 White controls, the mean plasma FXI was 21.5 μg mL−1 with standard deviation 12.1 μg mL−1. The mean level among the 248 Black controls was 21.1 μg mL−1 (SD = 8.1 μg mL−1). The mean of the logarithm of plasma FXI is not statistically significantly different for White and Black controls. On the other hand, the mean level of FXI was statistically significantly higher among Black cases (24.2 μg mL−1) compared with Black controls. Twenty-three (23.5%) cases had FXImeasurements above the 90th percentile of the control distribution (30.5 μg mL−1; odds ratio [adjusted for age, sex and body mass index] = 2.6; 95% CI, 1.4, 5.0).
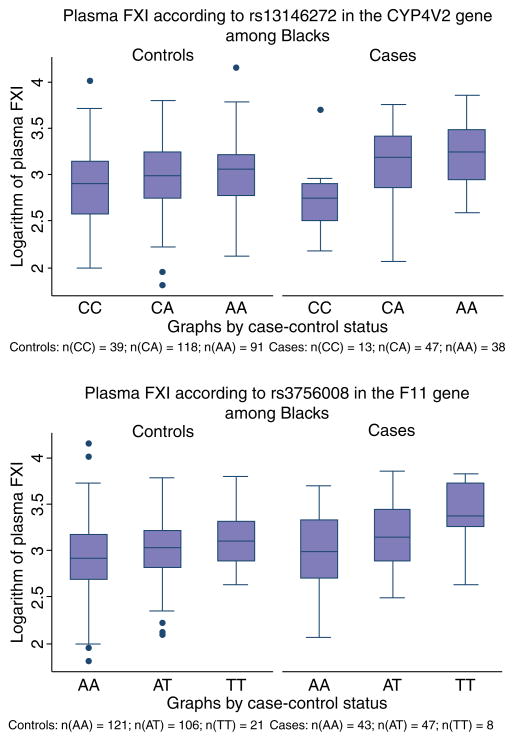
Among White controls only the genotypes corresponding to rs3087505 in the KLKB1 gene were associated with plasma FXI (P = 0.03; Table 3). Among Blacks, rs13146272 in the CYP4V2 gene and rs3756008 in the F11 gene were associated with plasma FXI. The associations were evident among both controls and cases and the associations were slightly stronger among cases (Table 3, Fig. 1).
Table 3.
Standardized regression coefficients for four single-nucleotide polymorphisms (SNPs) on chromosome 4 with the logarithm of plasma FXI for Whites and Blacks
| SNP | Controls
|
||
|---|---|---|---|
| Caucasions (n = 152) | Black (n = 248) | Black* (n = 346) | |
| rs13146272 | 0.075† (> 0.20)‡ | 0.131 (0.04) | 0.193 (0.0002) |
| rs3087505 | 0.174 (0.03) | 0.059 (> 0.20) | 0.074 (0.17) |
| rs2036914 | 0.042 (> 0.20) | 0.079 (> 0.20) | 0.089 (0.10) |
| rs3756008 | 0.075 (> 0.20) | 0.162 (0.01) | 0.198 (0.0002) |
Includes cases and controls using a regression model that also includes a term for case–control status.
Standardized coefficient from regression of log plasma FXI on the genotypes for that SNP.
Twotailed P value.
Fig. 1.
Box plot of plasma FXI by rs13146272 and rs3756008 among Blacks. Box edges mark the lower 25th percentile and the upper 75th percentile of the distribution. The upper whisker is X[75] + 1.5 * (X[75] − X[25]) and the lower whisker is X[25] − 1.5 * (X[75] − X[25]) + where X[i] is the ith percentile.
In a recent report of a genome-wide association study (GWAS), Tregouet et al. [10] evaluated SNPs rs2227589 (SERPINC1 gene), rs13146272 (CYP4V2 gene) and rs1613662 (GP6 gene) in three case–control studies. In Table 4 the findings from five verification case–control studies (including the three reported by Tregouet et al.) for the three SNPs identified by Bezemer et al. as statistically significantly associated with VTE are displayed. The overall association for the SERPINC1 SNP (OR = 1.15) is highly statistically significant (P = 0.004) and each study is consistent with the pooled overall OR (P value for a test of homogeneity > 0.20). The CYP4V2 SNP has an overall OR of 1.17 (P < 0.0001) and findings are consistent across studies (P for homogeneity > 0.20). A similar pattern is found for the GP6 SNP, with an overall OR of 1.14 (P = 0.002) and consistency across studies (P for homogeneity > 0.20).
Table 4.
Distribution of cases and controls, allele frequency and odds ratios with 95% confidence intervals for three single-nucleotide polymorphisms (SNPs) in five case–control studies
| Cases | Controls | OR | 95% CI | |
|---|---|---|---|---|
| SERPINC1 gene – rs2227589 | ||||
| MEGA2* | 1294† (0.12)‡ | 2836† (0.10)‡ | 1.29§ | 1.11, 1.49 |
| GWAS¶ | 419 (0.12) | 1228 (0.10) | 1.23 | 0.96, 1.57 |
| MARTHA¶ | 1150 (0.11) | 801 (0.11) | 1.00 | 0.82, 1.23 |
| FARIVE¶ | 607 (0.10) | 607 (0.10) | 1.00 | 0.77, 1.30 |
| GATE** | 544 (0.11) | 664 (0.11) | 1.07 | 0.83, 1.39 |
| OR†† = 1.15 (1.05, 1.26) | ||||
| CYP4V2 gene – rs13146272 | ||||
| MEGA2 | 1160 (0.69) | 2624 (0.64) | 1.24 | 1.12, 1.38 |
| GWAS | 419 (0.67) | 1228 (0.63) | 1.19 | 1.01, 1.40 |
| MARTHA | 1150 (0.64) | 801 (0.62) | 1.09 | 0.96, 1.24 |
| FARIVE | 607 (0.63) | 607 (0.61) | 1.09 | 0.92, 1.28 |
| GATE | 544 (0.66) | 661 (0.62) | 1.22 | 1.03, 1.44 |
| OR = 1.17 (1.10, 1.25) | ||||
| GP6 gene – rs1613662 | ||||
| MEGA2 | 1299 (0.84) | 2848 (0.82) | 1.14 | 1.01, 1.30 |
| GWAS | 419 (0.87) | 1228 (0.85) | 1.18 | 0.94, 1.48 |
| MARTHA | 1150 (0.86) | 801 (0.85) | 1.08 | 0.90, 1.30 |
| FARIVE | 607 (0.87) | 607 (0.84) | 1.27 | 1.01, 1.60 |
| GATE | 546 (0.85) | 663 (0.84) | 1.04 | 0.84, 1.30 |
| OR = 1.14 (1.05, 1.23) | ||||
From reference [1].
Number of subjects (n) in study.
Prevalence of the risk allele in 2N gametes (Table 1 defines risk allele).
Odds of risk allele in cases divided by the corresponding odds in control (2N gametes).
From reference [10].
The present study, White subjects only.
Summary odds ratio over the five studies.
Discussion
We replicated the findings of Bezemer et al. for rs13146272 in the CYP4V2 gene, for rs3087505 in the KLKB1 gene and for rs3756008 and rs2036914 in the F11 gene among Whites in the GATE study. We did not replicate the findings of Bezemer et al. for the SNPs in the SERPINC1 and GP6 genes. In multivariate analysis among Whites, only rs3756008 in F11 was statistically significantly associated with VTE. Among Blacks, we found statistically significant associations for rs2036914 in the F11 gene and for rs670659 in the RGS7 gene.
The SNPs in the region of the CYP4V2 (rs3756008 in F11, rs3087505 in KLKB1 and rs2036914 in F11) reside on genes that are involved in coagulation. The KLKB1 gene encodes for prekalikrein and the F11 gene encodes for FXI. The SERPINC1 gene encodes for antithrombin. The risk allele in the rs2227589 SNP in SERPINC1 has been associated with lower anticoagulant activity, increased plasma anti-FXa and increased antithrombin [11]. The CYP4V2 gene encodes for a protein that is not known to be related to thrombosis. However, the rs13146272 SNP in the CYP4V2 gene is in linkage disequilibrium with two SNPs in the F11 gene that have been related to VTE. It has been suggested that the association of rs13146272 with VTE and FXI levels is explained by the presence of the risk allele for rs13146272 on a common haplotype that includes the risk alleles for rs2036914 and rs2289252 in the F11 gene [12].
Our study findings with respect to plasma FXI among Whites are conflicting. With the exception of rs3087505 in the KLKB1 gene, we found little, or no, association between those SNPs that we found were related to case–control status and plasma FXI. These results conflict with those of Bezemer et al. We believe the reason for the discrepancy is that our study is underpowered among Whites. We used a different plasma FXI assay than did these investigators and the dispersion of the measurement among our subjects was considerably greater than was the corresponding dispersion in their study. Lack of power was not as much a problem among our Black subjects because we assayed twice as many controls among them. Among Blacks we found some persuasive positive associations between genotypes and plasma FXI (rs13146272 in CYP4V2 and rs3756008 in F11), although the association for rs2036914 in F11 (the SNP associated with case–control status) was weaker. Plasma FXI was elevated among Black cases compared with controls, as has been observed among Whites.
Among Whites, our findings for SNPs rs2227589 in SERPINC1 and rs1613662 in GP6 were null, but our study had little statistical power to detect an association with VTE for these SNPs. For rs2227589 the trend OR from the MEGA2 study was 1.29 and our study had about 50% power to detect an OR of this magnitude. However, at the upper 95% confidence limit for the MEGA2 trend OR (about 1.5) our study had about a 90% chance of detecting an OR this large. For rs1613662 in GP6 our study power is only about 60% even at the upper limit of the MEGA2 OR. The statistical power for the three SNPs (rs670659 in RGS7, rs2001490 in NR1I2 and rs1523127 in NAT8B) with false discovery rates < 0.20 in Bezemer et al. was below 50%. This is not surprising because the trendORfor each of these SNPs in MEGA2 is small, about 1.10. Thus, the absence of an effect for these SNPs in GATE, a smaller study than MEGA2, is not unanticipated.
We did post-hoc power calculations among Blacks using the prevalences of the nine risk alleles observed in this study and the trend OR for Causcasians in the MEGA2 study. The statistical power was highest (82%) for rs2036914 in F11, a SNP with an observed P < 0.05. The power for the three SNPs in Table 4 (rs2227589 in SERPINC1, rs13146272 in CYP4V2 and rs1613662 in GP6) was about 30%, 70% and 25%, respectively. At the upper 95% confidence limit for the MEGA2 trend OR, the power was about 70% for rs2227589 and rs1613662 while for rs13146272 it was about 95%. For rs3756008 in F11 the power was marginal (70% at the point estimate of the trend OR). The power was below 50% for rs3087505 in F11, rs1523127 in RGS7 and rs2001490 in NAT8B. Despite the low statistical power, we did find a positive effect for rs670659 in RGS7 in Blacks (P = 0.03, q = 0.09), an observation that, if verified, could prove important in understanding the genetic determinants of VTE in Blacks. Thus, the null findings in this study, especially among Blacks, must be interpreted with considerable caution because of the lack of statistical power for many of these SNPs.
Our meta-analysis indicates that available epidemiologic studies are consistent with the Bezemer et al. original report with respect to rs2227589, rs13146272 and rs1613662. We believe that the studies included in the meta-analysis make a persuasive case that these SNPs are related to VTE. TheGATE study provides strong support that rs13146272 and three other SNPs in its vicinity are risk factors for VTE among Whites. Although we found only weak associations for rs2227589 and rs1613662 among Whites in the GATE study, the meta-analysis indicates that the findings from the five epidemiologic studies are consistent within chance variation and hence the GATE findings add to the general literature through their inclusion in the meta-analysis.
Our study has other limitations in addition to low statistical power for some SNPs. Our VTE cases included only hospitalized patients. Persons with VTE treated as outpatients were not included. Hence, our findings may apply only to cases with disease requiring hospitalization. We also used controls from a clinic who may not provide an unbiased estimate of the population incidence of the genetic traits we evaluated. Furthermore, the participation rate of cases and controls was low [2]; about 40% of subjects asked to participate in the study refused, allowing for the introduction of bias in our estimates. However, we know of no reason why the incidence of the genetic traits studied here would differ for persons with VTE treated as in- or outpatients. Our controls were generally healthy and we believe do provide a reasonable estimate of the incidence of these genetic traits in the general population because the prevalence of the variant alleles among Whites in our study is comparable to that reported by Bezemer et al. in their study. Furthermore, we believe it unlikely that participation in the study is related to these genetic traits and therefore believe any selection bias is minimal. Thus, despite the limitations of our study, we believe that its findings are valid. We obtained plasma FXI measurements among a subset of subjects. We assayed mostly Blacks because little is known about the etiology of VTE among them. Our plasma FXI findings among Whites are of limited use because of small sample size and we did not assay White cases because we believed that the association between plasma FXI and VTE is well established. Our findings may not be comparable to those of Bezemer et al. because we included VTE cases with an underlying provocation, including cancer, while they excluded VTE cases with cancer. However, we analyzed our data after excluding 282 VTE cases with cancer and the results were essentially identical to those reported here.
We undertook this study to verify the Bezemer et al. findings among our White subjects and also evaluated the SNPs among our Black subjects. Thus, the motivation to evaluate these SNPs was based on observations on White Europeans and therefore it was not clear if these findings would pertain to Blacks. The observations among Blacks that rs2036914 in the F11 gene is a potential risk factor, that plasma FXI is associated with at least two of these SNPs, and that plasma FXI is higher among cases compared with controls, suggest a common genetic etiology, at least with respect to the F11 gene. In any event, we emphasize that the genetic etiology of VTE among Blacks is poorly understood and our work here was only a very limited evaluation of genetic determinants of VTE in Blacks because of limited statistical power for many of these SNPs. We plan additional, more thorough studies.
In conclusion, we were able to replicate the findings of Bezemer et al. for rs13146272 in the CYP4V2 gene and for three SNPs on two other genes in a region close by: rs3756008 and rs2036914 in F11 and rs3087505 in the KLKB1 gene. Among Whites, only rs3756008 in the F11 gene was related to VTE in a multivariate analysis. This is the first study that has evaluated these SNPs and plasma FXI among Blacks. Among Blacks, rs2036914 in the F11 gene was associated with VTE, as was plasma FXI. There is biologic credibility for a role of SNPs in the F11 gene in the etiology of VTE. Plasma FXI levels are related to VTE [1,3] and the risk alleles for rs2036914 and rs2289252 in F11 have been related to increased plasma FXI [1,12]. The positive association for rs670659 in the RGS7 gene among Blacks was not observed among Whites in our study, but rs670659 was associated with VTE in the Dutch studies [1]. Although we cannot rule out that the positive association for rs670659 is a chance finding among Blacks, our study suggests a potential role of the RGS7 gene in the etiology of VTE among Blacks, but this observation requires validation.
Acknowledgments
This work was supported by a grant from the CDC through the Associations of Schools of Public Health/CDC Cooperative Agreement mechanism and was carried out at Emory University.
H. Austin, W. C. Hooper and C. Lally designed the study and collected the data. W. C. Hooper and C. De Staercke were responsible for the laboratory work. Data management and analysis were carried out by C. Lally and H. Austin. All of the authors contributed to the writing, with H. Austin writing the first draft. F. R. Rosendaal and I.D. Bezemer identified the SNPs evaluated in this study, provided key scientific leadership, and contributed to the analysis and writing.
Footnotes
Disclosure of Conflict of Interests
The authors state that they have no conflict of interest.
References
- 1.Bezemer ID, Bare LA, Doggen CJ, Arellano AR, Tong C, Rowland CM, Catanese J, Young BA, Reitsma PH, Devlin JJ, Rosendaal FR. Gene variants associated with deep vein thrombosis. JAMA. 2008;299:1306–14. doi: 10.1001/jama.299.11.1306. [DOI] [PubMed] [Google Scholar]
- 2.Austin H, Key NS, Benson JM, Lally C, Dowling NF, Whitsett C, Hooper WC. Sickle cell trait and the risk of venous thromboembolism among blacks. Blood. 2007;110:908–12. doi: 10.1182/blood-2006-11-057604. [DOI] [PubMed] [Google Scholar]
- 3.Meijers JC, Tekelenburg WL, Bouma BN, Bertina RM, Rosendaal FR. High levels of coagulation factor XI as a risk factor for venous thrombosis. N Engl J Med. 2000;342:696–701. doi: 10.1056/NEJM200003093421004. [DOI] [PubMed] [Google Scholar]
- 4.Sham P. Statistics in Human Genetics. New York, NY: Oxford University Press Inc; 1998. [Google Scholar]
- 5.Devlin B, Risch N. A comparison of linkage disequilibrium measures of fine-scale mapping. Genomics. 1995;29:311–22. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- 6.Hosmer DW, Lemeshow S. Applied Logistic Regression. 2. New York, NY: Wiley and Sons Inc; 2000. [Google Scholar]
- 7.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–88. [Google Scholar]
- 8.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.StataCorp. Stata Statistical Software: Release 10. College Station. TX: Stata Corporation LP; 2007. [Google Scholar]
- 10.Tregouet DA, Heath S, Saut N, Biron-Andreani C, Schved JF, Pernod G, Galan P, Drouet L, Zelenika D, Juhan-Vague I, Alessi MC, Tiret L, Lathrop M, Emmerich J, Morange PE. Common susceptibility alleles are unlikely to contribute as strongly as the FV and ABO loci to VTE risk: results from a GWAS approach. Blood. 2009;113:5298–303. doi: 10.1182/blood-2008-11-190389. [DOI] [PubMed] [Google Scholar]
- 11.Anton AI, Teruel R, Corral J, Minano A, Martinez-Martinez I, Ordonez A, Vicente V, Sanchez-Vega B. Functional consequences of the prothrombotic SERPINC1 rs2227589 polymorphism on antithrombin levels. Haematologica. 2009;94:589–92. doi: 10.3324/haematol.2008.000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Bezemer ID, Rowland CM, Tong CH, Arellano AR, Catanese JJ, Devlin JJ, Reitsma PH, Bare LA, Rosendaal FR. Genetic variants associated with deep vein thrombosis: the F11 locus. J Thromb Haemost. 2009;7:1802–8. doi: 10.1111/j.1538-7836.2009.03544.x. [DOI] [PubMed] [Google Scholar]