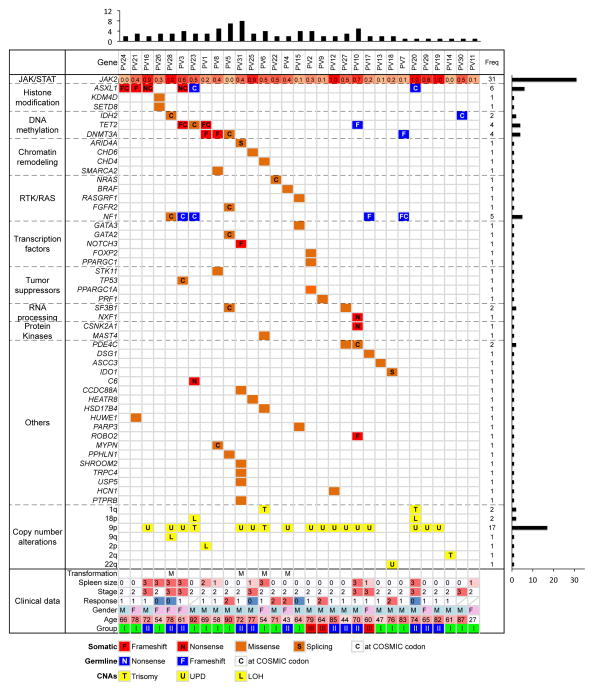
Figure 1. Summary of somatic mutations, germline variants and somatic DNA copy-number alterations identified in 31 PV cases.
Somatic mutations were shown in orange and red based on their mutation type and germline variants were shown in blue. For germline variants, only those frame-shift variants, nonsense variants or those variants reported in COSMIC database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/) were shown and only those genes with somatic mutations identified in the cohort were listed. The total number of somatic mutations identified per patient and gene was showed at the top and right, respectively. The allelic fraction of JAK2V617F was shown at the top row and color scaled. The group classification was based on the relationship of JAK2V617F and 9p aUPD. Subgroup I, patients harbored JAK2V617F in a heterozygous state without detectable 9p aUPD; Subgroup II, patients had JAK2V617F with an allelic fraction in direct proportion to the level of 9p aUPD; Subgroup III, patients harbored 9p aUPD at approximately twice the level of the JAK2V617F allelic burden. The clinical data were summarized at the bottom. CNAs, somatic DNA copy-number alterations: T, trisomy; L, hemizygous deletion; U, uniparental disomy. Transformation: M, myelofibrosis. Spleen size: 0, the size of spleen is not palpable; 1, < 3cm; 2, < 6cm; 3, > 6cm. Stage: 1, low; 2, intermediate; 3, high. Response to therapy: 0, no response; 1, partial response; 2, complete response. Gender: M, male; F, female.