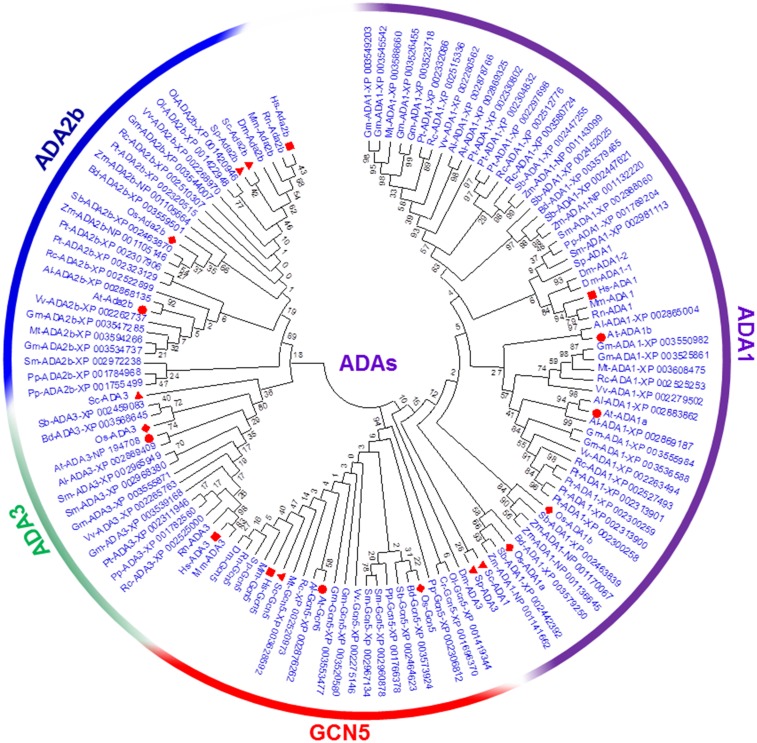
Fig 2. Phylogenetic relationship of the ADAs protein of the SAGA complex.
ADAs protein sequences were used from At, Arabidopsis thaliana (red circle); Dm, Drosophila melongaster; Hs, Homo sapiens (red square); Os, Oryza sativa (red diamond shape); Mm, Mus musculus; Rs, Rattus norvegicus; Sc, Saccharomyces cerevisiae (red triangle); Sp, Schizosaccharomyces pombe; Zm, Zea mays; Rc, Ricinus communis; Pt, Populus trichocarpa; Vv, Vitis vinifera; Al, Arabidopsis lyrta; Mt, Medicago truncatula; Bd, Brachypodium distachyon; Sb, Sorgum bicolor; Sm, Selaginella moellendorffii; Pp, Physcomitrella patens; Cr, Chlamydomonas reinhardtii and Ol, Ostreococcus lucimarinus. Phylogeny reconstruction was analysed by neighbour-joining statistical method. Test of phylogeny was analysed by the bootstrap method (1,000 replicates).