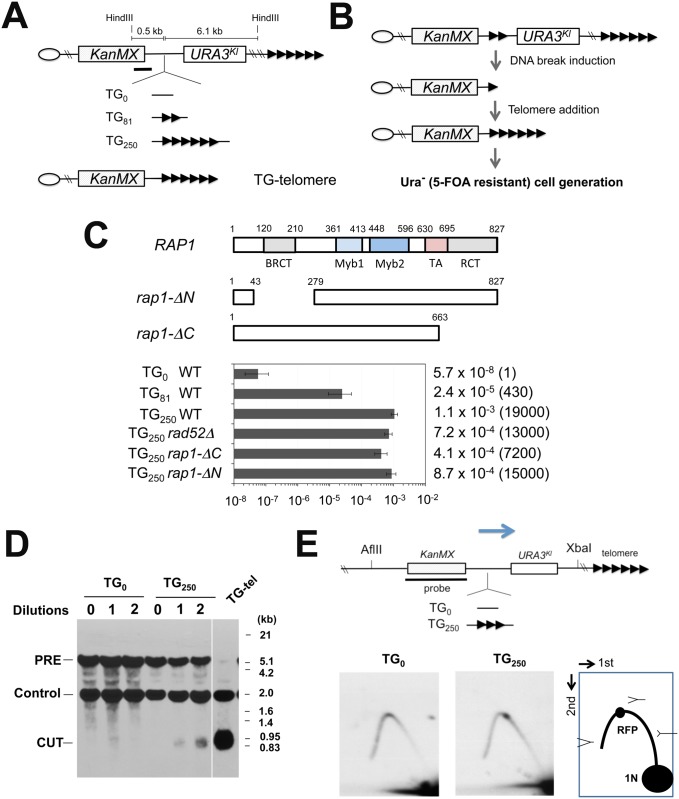
Fig 1. Chromosome truncation at internal TG repeats.
(A) Schematic of the 81 bp TG (TG81) or 250 bp TG (TG250) repeat at the ADH4 locus on chromosome VII-L. The ADH4 locus was replaced with a DNA fragment containing the KanMX gene and the URA3 Kl gene. The TG81 or TG250 sequence was introduced between KanMX and URA3 Kl. The cassette with no TG sequence was called as TG0. The centromere is shown as a circle on the left (CEN) and the telomere is shown as repetitive arrows on the right. The black line indicates a hybridization probe for Southern blot (Fig 1C). TG-telomere cells contain a telomere at the position where the TG250 repeat is inserted (See S1 Fig for more details). (B) Schematic of experimental protocol to detect URA3 Kl marker loss after telomere addition at the TG repeat sequences. (C) Effect of TG81 or TG250 insertion on URA3 marker loss. Cells containing the TG0, TG81 or TG250 cassette were first maintained in medium selective for URA3 and then transferred to non-selective medium. Saturated cultures were diluted and spread out on 5-FOA plates to determine the rate of URA3 Kl marker loss. URA3 Kl marker loss rate per generation was quantified through fluctuation analysis. Error bars represent 95% confidence intervals. Number in parentheses indicates rate relative to cells containing the TG0 cassette. Rap1 contains a BRCT domain, two Myb domains, a transcription activation (TA) domain and a C-terminal RCT domain. The rap1-ΔN and the rap1-ΔC mutation lack the BRCT and the RCT domain, respectively. (D) Effect of TG250 insertion on chromosome truncation. Cells containing the TG0 or TG250 cassette were first maintained in medium selective for URA3 and then diluted 1000-fold in non-selective medium to allow cells to grow for one day (10 generations) and aliquots were collected for genomic DNA preparation. This cycle was repeated again. DNA was digested with HindIII and analyzed by Southern blot using the probe shown in A. DNA breakage at TG250 generates a 0.7 kb fragment (marked with CUT) from the 6.9 kb fragment (PRE). The probe also detects a 1.8 kb HindIII fragment (Control) from the SMC2 locus on chromosome VI. TG-telomere (TG-tel) serves as a control to detect DSB induction at the TG250 sequence. (E) Effect of TG250 insertion on replication fork pausing. Cells containing the TG0 or TG250 cassette were first maintained in medium selective for URA3 and then transferred to non-selective medium to allow cells to undergo cell division for 4 hr. CsCl gradient purified DNA was digested with AflII and XbaI and analyzed by two-dimensional gel electrophoresis using the indicated probe. The probe detects a 6.3 kb AflII-XbaI fragment. The TG250 repeat locates 3.0 kb from the AflII site and 2.9 kb from the XbaI site. RFP represents replication fork pausing. The arrow indicates the direction of replication fork movement. There is a highly active replication origin 40 kb proximal to the repeat insert site (the ADH4 locus) on chromosome VII [82].