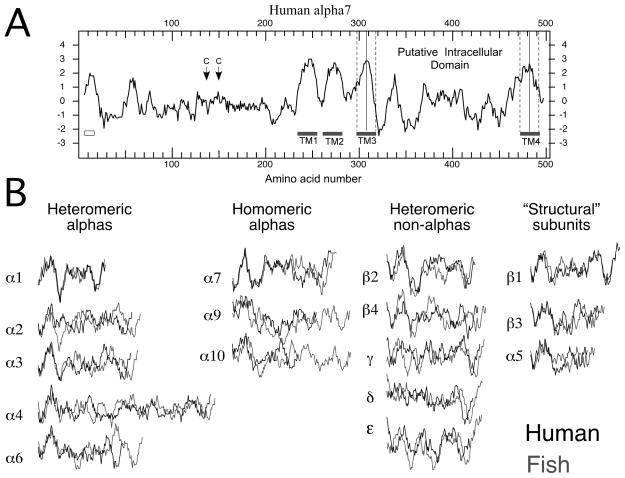
Figure 2.
A) Kyte-Doolittle hydrophobicity plot of human α7 from DNA Strider (version 1.4f17 CEA France). Plotted are the average hydrophobicity scores of 13 amino acids in a rolling window. Long stretches of amino acids with positive scores are likely to be lipid associated. The amino terminal signal sequence is indicated by the box under the plot on the left. Position of the two cysteine residues which define the signature Cys-loop are indicated by the arrows. Gray bars indicate the regions associated with the transmembrane domains. The dashed lines indicate how the putative intracellular domains were selected as delimited by 10 amino acids from the hydrophobic peaks of TM3 and TM4. B) The putative intracellular domains of all the nAChR excised from the Kyte-Doolittle hydrophobicity plots for each subunit. The profiles in black are from human sequences, and those in gray are from zebrafish sequences. Vertical alignments are based on the Kyte-Doolittle scores, and the horizontal alignments are based on the putative amino terminals of the respective intracellular domains determined as described for panel A.