Abstract
Genome-wide association studies implicate the MIR137HG risk variant rs1625579 (MIR137HGrv) within the host gene for microRNA-137 as a potential regulator of schizophrenia susceptibility. We examined the influence of MIR137HGrv genotype on 17 subcortical and callosal volumes in a large sample of individuals with schizophrenia and healthy controls (n=841). Although the volumes were overall reduced relative to healthy controls, for individuals with schizophrenia the homozygous MIR137HGrv risk genotype was associated with attenuated reduction of mid-posterior corpus callosum volume (p=0.001), along with trend-level effects in the adjacent central and posterior corpus callosum. These findings are unique in the literature and remain robust after analysis in ethnically homogenous and single-scanner subsets of the larger sample. Thus, our study suggests that the mechanisms whereby MIR137HGrv works to increase schizophrenia risk are not those that generate the corpus callosum volume reductions commonly found in the disorder.
Keywords: microRNA-137, MIR137, schizophrenia, subcortical volume, corpus callosum, multi-site
1. Introduction
Recent research implicates a variant within the host gene (MIR137HG) for microRNA-137 (miR-137) as a contributing factor to the genetic regulation of schizophrenia [1]. The single nucleotide polymorphism (SNP) rs1625579 (MIR137HGrv) showed the strongest association with the illness (p=1.6×10−11) in a large-scale genome-wide association study (GWAS) of the disorder [2], the results of which were recently replicated in an independent dataset [3]. Although MIR137HGrv is particularly relevant to GWAS, other nearby SNPs relating to schizophrenia have been demonstrated to decrease miR-137 expression via altering the secondary structure of the primary transcript (4). Individuals with the MIR137HGrv homozygous schizophrenia-risk genotype express lower levels of miR-137, indicating that the risk polymorphism likely plays a role in regulating microRNA activity and subsequent expression of target genes [5]. While the association of MIR137HGrv with schizophrenia is intriguing, its specific contribution to endophenotypes of the disorder remains unclear. The goal of the present analysis is to elucidate the influence of the MIR137HGrv risk genotype on structural brain variation in schizophrenia.
Subcortical volumes are abnormal in individuals with schizophrenia, with the largest effect sizes belonging to an increase in the lateral ventricles and to reductions in the hippocampus and thalamus [8; 9]. A recent study by Lett and colleagues [10] assessed the relationship between MIR137HGrv genotype and select subcortical brain volumes, discovering smaller hippocampal volumes and larger lateral ventricles in individuals with schizophrenia but in not healthy controls with a homozygous MIR137HGrv risk allele genotype. However, the analysis examined only three brain regions and included a limited sample size (n=213). In genetic analyses, the importance of replication cannot be overstated, given the small effect sizes and the possibility of false positives. Therefore, further investigation is required to verify these findings and discover the extent to which MIR137HGrv may influence other regions across the brain affected in schizophrenia, including the thalamus, amygdala, and cerebellum [8]. The corpus callosum is also affected in schizophrenia, with different patterns of effect for different subregions of the structure [11].
We present an analysis of the influence of MIR137HGrv genotype on volumes of 12 subcortical structures and 5 corpus callosum measures in 841 schizophrenia patients and controls from an aggregated dataset.
2. Methods
2.1 Data collection
2.1.1 Participants
Analyses were conducted on 841 participants from six independent subsamples and nine imaging sites. All studies were conducted with local IRB approval and all subjects provided informed consent. Table 1 shows the distribution of age, sex, diagnosis, and MRI scanner type for each dataset.
Table 1.
Dataset demographic information. F, female; M, male; SZ, schizophrenia patient; HC, healthy control.
| Dataset | N | Sex (F/M) | Age (mean/SD) | Diagnosis (SZ/HC) | Scanner type |
|---|---|---|---|---|---|
| MCIC (4 sites) | 211 | 70/141 | 34.04/10.88 | 98/113 | GE 1.5T, Siemens 1.5T (2), Siemens 3T |
| NU | 130 | 48/82 | 34.28/12.96 | 69/61 | Siemens 1.5T |
| COBRE-1 | 68 | 13/55 | 37.03/13.62 | 32/36 | Siemens 3T |
| COBRE-2 | 87 | 27/60 | 38.29/13.06 | 50/37 | Siemens 3T |
| Dublin | 247 | 133/114 | 31.58/12.21 | 43/205 | Philips Intera Achieva 3T |
| Galway | 99 | 38/61 | 35.87/9.90 | 67/32 | Philips Intera Achieva 1.5T |
| TOTAL | 841 | 331/520 | 34.2/12.19 | 362/490 |
2.2 Genotyping
Genetic data were derived from DNA extracted from participant blood and saliva samples (see Supplementary Material). Table 2 outlines the relative genotype frequencies and ethnicity distribution of subjects across datasets. The major T allele is considered the MIR137HGrv schizophrenia risk allele [2].
Table 2.
Proportion of each dataset by ethnicity and minor allele frequency. Numbers in parentheses indicate the proportion of G allele carriers of a particular ethnicity group in a particular subsample. GG and GT genotype individuals were collapsed into one group for analysis due to low frequency of the G allele.
| Dataset | N | Caucasian (G) | Hispanic/East Indian (G) | African American (G) | Total G allele frequency |
|---|---|---|---|---|---|
| MCIC1 | 46 | 0.72 (0.12) | 0 | 0.28 (0.19) | 0.15 |
| MCIC2 | 82 | 1 (0.18) | 0 | 0 | 0.18 |
| MCIC3 | 43 | 0.77 (0.18) | 0 | 0.23 (0.20) | 0.19 |
| MCIC4 | 39 | 0.46 (0.14) | 0.54 (0.12) | 0 | 0.13 |
| NU | 130 | 0.59 (0.16) | 0 | 0.41 (0.3) | 0.22 |
| COBRE1 | 68 | 0.63 (0.20) | 0.37 (0.16) | 0 | 0.18 |
| COBRE2 | 87 | 0.37 (0.26) | 0.63 (0.10) | 0 | 0.16 |
| Dublin | 247 | 1 (0.16) | 0 | 0 | 0.16 |
| Galway | 99 | 1 (0.18) | 0 | 0 | 0.18 |
| Total | 851 | 0.79 (0.17) | 0.13 (0.12) | 0.09 (0.27) | 0.20 |
2.3 Magnetic Resonance Image (MRI) acquisition
MRI scans were collected from multiple scanners including a GE 3T, Philips Intera Achieva, Siemens 1.5T, Siemens 3T, and Philips 3T scanner. Details on imaging procedures are available in the Supplementary Material.
2.4 Subcortical segmentation of MRI data
For multi-site datasets in general, the characteristics of subcortical volumes have been validated through standardized procedures and meta-analyses conducted by the ENIGMA Consortium [12]. Subcortical volumetric segmentation of structural MRI scans was performed using FreeSurfer software (v.5.1.0 for Dublin dataset, v.5.3.0 for the remainder; www.surfer.nmr.mgh.harvard.edu). After outlier identification and visual inspection to verify segmentation accuracy, we included in our analysis the lateral ventricles, inferior lateral ventricles, putamen, globus pallidus, hippocampus, thalamus, caudate nucleus, nucleus accumbens, amygdala, cerebellar cortex, and cerebellar white matter, as well as the corpus callosum.
The FreeSurfer processing pipeline further segmented the corpus callosum into five segments of equal length on the sagittal plane: the anterior, mid-anterior, central, mid-posterior, and posterior regions. This parcellation closely approximates widely accepted designations of corpus callosum subregions [13]. Comparison of the accuracy of FreeSurfer automated segmentation to manual tracing shows high correlation and comparable accuracy between both methods [14].
2.5 Correcting heterogeneity in the data
Ethnic stratification was characterized in our datasets by the ENIGMA Consortium’s multi-dimensional scaling (MDS) procedure on genome-wide scan data when available (Fig. S1) [15]. For the European Dublin and Galway datasets, genome-wide scan data were unavailable. All participants self-reported as Caucasian, and were categorized as such with confidence given the overwhelming prevalence of Caucasian ethnicity in their population. All Asian and mixed ethnicity participants were excluded due to low sample size (total n=20), for a final sample size of 841 (Table 2).
2.6 Analytic approach
Due to low frequency of the GG genotype, GG and GT participants were combined (n=274) for comparison to the ‘homozygous-risk’ TT genotype participants (n=567).
The influence of MIR137HGrv genotype on subcortical volumes was examined via repeated measures ANCOVAs [10] and a MANCOVA (IBM SPSS Statistics, v.21). Dummy-coded genotype, diagnosis, categorical ethnicity, and imaging site served as between-group factors, and age, sex, and FreeSurfer-estimated total intracranial volume served as covariates. Model selection via backward elimination for significant effects identified a final model including all main effects and the site by sex, site by diagnosis, and genotype-by-diagnosis interaction terms. The genotype-by-diagnosis interaction term did not survive backward elimination but remained in the model due to previously discovered incongruent effects of MIR137HGrv genotype in schizophrenia patient and healthy control populations [10].
We applied the MANCOVA model to the five bilateral corpus callosum regions. These regions were assessed separately given their disparate associations with schizophrenia in the literature [16; 17]. The left and right hemisphere volumes for the twelve subcortical structures were analyzed using repeated measures ANCOVA. Raw p-values underwent Bonferroni correction for five or twelve multiple comparisons (i.e. p/5 and p/12), for the MANCOVA and repeated measures ANCOVAs, respectively.
2.7 Subsample analyses
To further characterize any effect of ethnicity on our results, the complete analysis was performed on a Caucasian-only subsample of the data (n=663) for comparison to the mixed-ethnicity results.
Given the variety of scanning sites, the complete analysis was also performed on each dataset separately for comparison to the aggregated sample.
3. Results
3.1 Effects of diagnosis on brain volume
Patients demonstrated significantly different mean volumes from controls in several regions (Table S1). Notably, patients showed a significant decrease in total corpus callosum volume, and a significant increase in total lateral ventricle volume.
3.2 Interaction of MIR137HGrv genotype and diagnosis
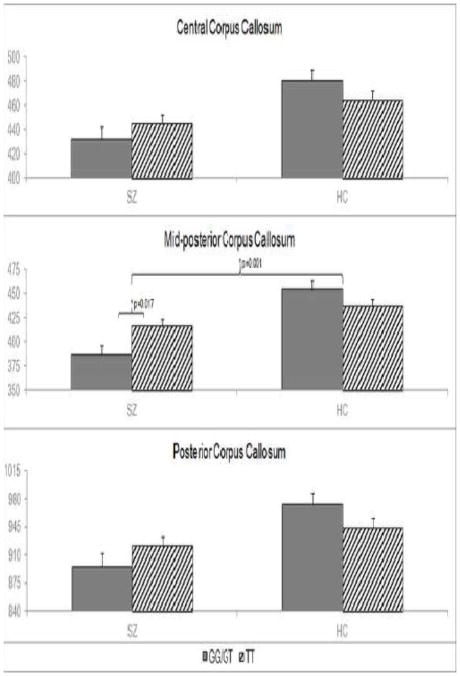
The corpus callosum MANCOVA revealed a genotype-by-diagnosis interaction in the mid-posterior corpus callosum (F1,808=11.97, p=0.001) that remained significant after Bonferroni correction (see Figure 1). There were also genotype-by-diagnosis interaction effects on the posterior corpus callosum (F1,808=5.94, p=0.015) and central corpus callosum (F1,808=3.82, p=0.051) that did not survive Bonferroni correction. The overall effect of genotype-by-diagnosis on the omnibus MANCOVA was significant as well (F5,804=2.88, p=0.014).
Figure 1.
Effect of genotype by diagnosis interaction on corpus callosum volumes.
A MANCOVA yielded an effect of MIR137 genotype by diagnosis interaction on the central (p=0.051), mid-posterior (p=0.001), and posterior corpus callosum (p=0.015). Post hoc analyses determined that in patients only, mean mid-posterior corpus callosum volume is significantly greater for TT than GG/GT carriers (p=0.017). Effects remaining significant after Bonferroni correction are marked with stars. Mean volume of patients and controls differed significantly in all three regions (p< 0.001).
The post-hoc pairwise comparison revealed that mid-posterior corpus callosum volume was significantly larger in TT than GG/GT carriers with schizophrenia (F1,804=4.21, pcorrected= 0.007), but there was no significant effect of genotype in controls. The mean volume of TT carriers with schizophrenia was significantly smaller than the mean volume of all controls (F1,835=10.59, p=0.001).
The repeated measures MANCOVAs did not yield a significant effect of genotype or genotype-by-diagnosis interaction on any bilateral region after Bonferroni correction. The direction of diagnosis effects between genotype groups on other volumes is illustrated in Figure S2.
3.3 Subsample analyses to address possible data confounds
3.3.1 Caucasian subset analysis
In the separate analysis conducted using only Caucasian subjects, a trend-level significant effect of genotype-by-diagnosis interaction was found on the central corpus callosum (F1,662=3.63, p=.057) and a significant effect in the mid-posterior corpus callosum (F1,662=8.64, p=.003). The direction of the genotype-by-diagnosis interaction in both regions was in accordance with those found in the full dataset: TT genotype patients showed greater volume than GG/GT counterparts, with the opposite or no effect in controls.
3.3.2 Consistency over individual datasets
We performed the MANCOVA and repeated measures ANCOVA analyses on each subsample separately. In the mid-posterior corpus callosum, all sites except one displayed a larger mean volume in TT patients than GG/GT patients. The subsamples generally displayed the same direction of genotype-by-diagnosis interaction effect consistent with whole dataset results. All of the subsamples’ results are shown in Supplemental Material Figure S3, along with the overall means.
4. Discussion
4.1 Effects of MIR137HGrv on subcortical volumes
This study assessed the relationship between MIR137HG SNP rs1625579 (MIR137HGrv) genotype and subcortical volumes in schizophrenia using a large, ethnically heterogeneous dataset. As would be expected from the association between MIR137HGrv and schizophrenia, our finding is significant in patients only. Given that MIR137HGrv confers risk for schizophrenia [2], and that the mean corpus callosum volume is reduced in the illness [16; 17], we expected that the MIR137HGrv homozygous risk genotype (TT) would lead to a reduced volume either in patients or both groups; but this is not the case. Rather, our findings suggest a counter-intuitive role of the variant in corpus callosum volume. In the mid-posterior corpus callosum, the homozygous risk allele genotype (TT) correlated with greater regional volume compared to GG/GT genotype, but in patients only. Similar though weaker effects were found in the adjacent posterior and central corpus callosum. The relationship between homozygous risk allele genotype and the relatively enlarged regional volume indicates that in patients, MIR137HGrv risk is associated with lesser volumetric reduction in the central and posterior regions of the corpus callosum. These results remained largely intact at trend-level significance for the site- and Caucasian-specific subsets of the data. Thus, our study suggests that the mechanisms whereby MIR137HGrv works to increase schizophrenia risk are not those that generate the corpus callosum volume reductions commonly found in the disorder.
4.2 MIR137HGrv shows no effect on hippocampus and lateral ventricles in larger sample
Our results differed considerably from the ventricular and hippocampal effects found in the Lett et al. [10] prior analysis of MIR137HGrv genotype on subcortical volumes. In contrast to their previous Caucasian-only analysis, our study uses a much larger dataset. Although the full sample was heterogeneous with respect to ethnicity, the Caucasian subset of our data displayed the same results at trend-level significance. Furthermore, we did not replicate the previous study’s ventricular or hippocampal findings even in the Caucasian subset, suggesting that our enhanced sample size likely improved the resolution of our findings (Fig S3). Our findings are supportive of Rose et al. 2014 who also found no effect of the SNP on hippocampal volume in a sample of both patients and controls [18].
4.3 The corpus callosum in schizophrenia
Abnormalities in the corpus callosum, the largest and most uniformly organized white matter structure in the human brain, are widely speculated to underlie disrupted neuronal connectivity in schizophrenia. While numerous structural MRI studies have identified reductions in corpus callosum size in individuals with schizophrenia, as we have here, there remains uncertainty in the degree to which the anterior, posterior, midbody (encompassing the mid-posterior and central segments), and total corpus callosum each contribute to disease presentation [19; 20]. Through manual tracing with a similar equal-lengths approach as FreeSurfer automated segmentation, Mitelman et al. 2009 identified increased area of only the posterior mid-body corpus callosum in patients compared to controls [11], in line with our direction of effects. Furthermore, several studies have found in the posterior midbody reduced regional fractional anisotropy [21] and in some cases increased mean diffusivity [22; 23], markers of diminished white matter organization potentially reflecting reduced regional axonal density in patients. The posterior corpus callosum develops independently from the anterior segments, instead developing earlier along with the hippocampal commissure [24], and may thus be influenced by hippocampal growth patterns.
4.4 MIR137HGrv and callosal development and function
Evidence suggests that genes targeted by miR-137 act to regulate axonal guidance and myelination mechanisms responsible for generating the various components of the corpus callosum and other white matter tracts in the brain. A bioinformatics analysis of predicted and validated miR-137 target genes revealed a significant enrichment of these genes within the axonal guidance canonical pathway [1]. One target, ZNF804A, is a well-known schizophrenia candidate gene [25] that has been linked to reduced integrity of the corpus callosum [26], although its relationship to white matter integrity has varied across studies [27; 28]. Since many other miR-137 target genes are also schizophrenia risk genes [1], the role of these genes in white matter development and morphology lends support to our finding of MIR137HGrv genotype-by-diagnosis—rather than just genotype—effects on corpus callosum volume.
4.5 Alternative explanations of MIR137HGrv and schizophrenia risk
Our results indicate that MIR137HGrv risk does not relate directly to subcortical volumetric measures with the exception of the posterior corpus callosum, and are in keeping with other recent research that also did not report an effect of MIR137HGrv genotype on subcortical brain volumes in either healthy controls or patients [17]. The effect of MIR137HGrv may more strongly manifest through other measures of brain function. For example, dysfunctional connectivity between the dorsolateral prefrontal cortex (DLPFC) and hippocampus is a well-established phenotype of schizophrenia patients, and studies in healthy controls [29] as well as controls and patients [30] report that MIR137HGrv homozygous risk individuals show increased DLPFC-hippocampus connectivity, greater deficits in measures of episodic memory and attention span [31], and fail to show the relationship between DLPFC-hippocampal connectivity and memory performance that the non-risk allele subjects do [30]. These results suggest that perhaps the action of MIR137HGrv is not generally on brain structure or volumes but on the functional relationship between the frontal cortex and other regions in cognitive processing. Further research into connectivity particularly via the posterior corpus callosum may provide more insight into the relationship between MIR137HGrv and schizophrenia.
Alternatively, the effects of miR-137 on risk for schizophrenia may be mediated through multiple mechanisms. Several variants in the miR-137 gene have been associated with disrupted miR-137 expression in association with schizophrenia (4), suggesting it may affect a number of separate schizophrenia endophenotypes.
4.6 Study limitations
We considered whether aberrant datasets could have contributed to our unusual findings. Diagnostic group trends in our sample behaved as expected according to established volumetric differences in the literature [7], indicating that the observed genotype-by-diagnosis effects were not driven by abnormal group differences between patients and healthy controls (Fig. S2).
The benefits of the multi-site nature of our study outweigh the limitations of the approach, and while it is a possible confound to our genotype-by-diagnosis interaction effects, we did not identify a significant interaction between site, genotype, and diagnosis. The direction of our significant results remained relatively intact but underpowered for statistical significance when analyzing each of our six subdatasets individually (Fig. S3). Thus, genotype-by-diagnosis effects are relatively consistent across the samples in this dataset and suggest that our whole dataset results are likely to be replicated in other independent samples.
An important limitation of our study was the use of patient participants who were all on unknown medication regimens at the time of the MRI scan. There are demonstrated medication effects on basal ganglia volumes [33; 34], but meta-analysis suggests that total white matter loss is reduced over time at a comparable rate in medicated and antipsychotic-naïve patients [7]. Thus, volumetric differences in white matter may exist at disease onset without aggravation by antipsychotic treatment.
5 Conclusion
In summary, we present promising evidence that the MIR137HGrv homozygous risk genotype relates to less severe volumetric reduction of the midbody and posterior corpus callosum in schizophrenia patients. Thus, it is unlikely that the variant mediates enhanced risk for schizophrenia by exacerbating the corpus callosum volume loss found in the illness. The role of MIR137HGrv in schizophrenia remains an area of active research, and future studies can further the investigation by examining the integrated effects of the variant and miR-137 target genes.
Supplementary Material
Highlights.
We examine how the microRNA-137-associated variant rs1625579 affects subcortical volumes in schizophrenia.
In patients, the homozygous risk genotype confers lesser reduction of callosal volume.
Results are robust after considering possible multi-site and ethnicity confounds.
Results suggest variant does not mediate schizophrenia risk through corpus callosum volume reduction.
Acknowledgments
The authors gratefully acknowledge Jill Fries for her assistance with executing the FreeSurfer segmentation on the COBRE1, COBRE2, MCIC, and NU datasets. We also thank Dr. Ronald Yeo for his guidance with the early development of this manuscript.
This work was supported by the National Institutes of Health (grant numbers 5R01MH094524-03 to VDC and JAT, R01MH084898-01A1 to JRB, P50 MH071616, R01MH056584, 1R01 MH084803 to LW, and 1U01 MH097435 to LW and JAT); the United States Department of Energy (grant number DE-FG02-99ER62764); the Science Foundation Ireland (grand number SFI 12.IP.1359); the Irish Health Research Board (grant numbers HRA_POR/2011/100, HRA_POR/2012/54); and the Wellcome Trust (grant number 072894/2/03/Z).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Veena S. Patel, Email: vpatel@mrn.org.
Sinead Kelly, Email: kellys37@tcd.ie.
Carrie Wright, Email: clwright@salud.unm.edu.
Cota Navin Gupta, Email: ncota@mrn.org.
Alejandro Arias-Vasquez, Email: alejandro.ariasvasquez@radboudumc.nl.
Nora Perrone-Bizzozero, Email: nbizzozero@salud.unm.edu.
Stefan Ehrlich, Email: stefan.erhlich@uniklinikum-dresden.de.
Lei Wang, Email: leiwang1@northwestern.edu.
Juan R. Bustillo, Email: jbustillo@salud.unm.edu.
Derek Morris, Email: derek.morris@nuigalway.ie.
Aiden Corvin, Email: acorvin@tcd.ie.
Dara M. Cannon, Email: dara.cannon@nuigalway.ie.
Colm McDonald, Email: colm.mcdonald@nuigalway.ie.
Gary Donohoe, Email: gary.donohoe@nuigalway.ie.
Vince D. Calhoun, Email: vcalhoun@mrn.org.
Jessica A. Turner, Email: jturner@mrn.org.
References
- 1.Wright C, Turner JA, Calhoun VD, Perrone-Bizzozero N. Potential impact of miR-137 and its targets in schizophrenia. Front Genet. 2013;4:58. doi: 10.3389/fgene.2013.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan F, Zhang B, Yan T, Li L, Liu F, Li T, et al. MIR137 gene and target gene CACNA1C of miR-137 contribute to schizophrenia susceptibility in Han Chinese. Schizophr Res. 2014;152:1. doi: 10.1016/j.schres.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Strazisar M, Cammaerts S, van der Ven K, Forero DA, Lenaerts AS, Nordin A, et al. MIR137 variants identified in psychiatric patients affect synaptogenesis and neuronal transmission gene sets. Mol Psy. 2015;20:472–481. doi: 10.1038/mp.2014.53. [DOI] [PubMed] [Google Scholar]
- 5.Guella I, Sequeira A, Rollins B, Morgan L, Torri F, van Erp TGM, et al. Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex. J Psychiatr Res. 2013;47:9. doi: 10.1016/j.jpsychires.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Bio. 2010;189:1. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:6. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haijma SV, Van Haren N, Cahn W, Koolschijn PC, Hulshoff Pol HE, Kahn RS. Brain volumes in schizophrenia: a meta-analysis in over 18000 subjects. Schizophr Bull. 2013;39:5. doi: 10.1093/schbul/sbs118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lett TA, Chakavarty MM, Felsky D, Brandl EJ, Tiwari AK, Goncalves VF, et al. The genome-wide supported microRNA-137 variant predicts phenotypic heterogeneity within schizophrenia. Mol Psychiatry. 2013;18:4. doi: 10.1038/mp.2013.17. [DOI] [PubMed] [Google Scholar]
- 11.Mitelman SA, Nikiforova YK, Canfield EL, Hazlett EA, Brickman AM, Shihabuddin L, Buchsbaum MS. A longitudinal study of the corpus callosum in chronic schizophrenia. Schizophr Res. 2009;114:144–53. doi: 10.1016/j.schres.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, et al. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014;8:2. doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofer S, Frahm J. Topography of the human corpus callosum revisited-comprehensive fiber tractography using diffusion tensor magnetic resonance imaging. Neuroimage. 2006;32:989–94. doi: 10.1016/j.neuroimage.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 14.Clerx L, Gronenschild HB, Echavarri C, Verhey F, Aalten P, Jacobs HIL. Can Free Surfer compete with manual volumetric measurements in Alzheimer’s disease? Curr Alzheimer Res. doi: 10.2174/1567205012666150324174813. epub. [DOI] [PubMed] [Google Scholar]
- 15.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Edmiston Kale E, Chen K, Tang Y, Ouyang X, Jiang Y, et al. A comparative diffusion tensor imaging study of corpus callosum subregion integrity in bipolar disorder and schizophrenia. Psychiatry Res. 2014;221:1. doi: 10.1016/j.pscychresns.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Walterfang M, Wood AG, Reutens DC, Wood SJ, Chen J, Velakoulis D, McGorry PD, Pantelis C. Morphology of the corpus callosum at different stages of schizophrenia: cross-sectional study in first-episode and chronic illness. Br J Psychiatry. 2008;192:6. doi: 10.1192/bjp.bp.107.041251. [DOI] [PubMed] [Google Scholar]
- 18.Rose EJ, Morris DW, Fahey C, Cannon D, McDonald C, Scanlon C, et al. The miR-137 schizophrenia susceptibility variant rs1625579 does not predict variability in brain volume in a sample of schizophrenic patients and healthy individuals. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:467–71. doi: 10.1002/ajmg.b.32249. [DOI] [PubMed] [Google Scholar]
- 19.Gasparotti R, Valsecchi P, Carletti F, Galluzzo A, Liserre R, Cesana B, et al. Reduced fractional anisotropy of corpus callosum in first-contact, antipsychotic drug-naïve patients with schizophrenia. Schizophr Res. 2009;108:1–3. doi: 10.1016/j.schres.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Mitelman SA, Torosian Y, Newmark RE, Schneiderman JS, Chu KW, Brickman AM. Internal capsule, corpus callosum and long associative fibers in good and poor outcome schizophrenia: a diffusion tensor imaging survey. Schizophr Res. 2007;92:1–3. doi: 10.1016/j.schres.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 21.Ellison-Wright I, Nathan PJ, Bullmore ET, Zaman R, Dudas RB, Agius M, et al. Distribution of tract deficits in schizophrenia. BMC Psychiatry. 2014;14:99. doi: 10.1186/1471-244X-14-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holleran L, Ahmed M, Anderson-Schmidt H, McFarland J, Emsell L, Leemans A, et al. Altered interhemispheric and temporal lobe white matter microstructural organization in severe chronic schizophrenia. Neuropsychopharmacology. 2014;39:944–54. doi: 10.1038/npp.2013.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rotarska-Jagiela A, Schonmeyer R, Oertel V, Haenschel C, Vogeley K, Linden DE. The corpus callosum in schizophrenia-volume and connectivity changes affect specific regions. Neuroimage. 2008;39:1522–32. doi: 10.1016/j.neuroimage.2007.10.063. [DOI] [PubMed] [Google Scholar]
- 24.Raybaud C. The corpus callosum, the other great forebrain commissures, and the septum pellucidum: anatomy, development, and malformation. Neuroradiology. 2010;52:447–77. doi: 10.1007/s00234-010-0696-3. [DOI] [PubMed] [Google Scholar]
- 25.O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:9. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 26.Ikuta T, Peters BD, Guha S, John M, Karlsgodt KH, Lencz T, et al. A schizophrenia risk gene, ZNF804A, is associated with brain white matter microstructure. Schizophr Res. 2014;155:1–3. doi: 10.1016/j.schres.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voineskos AN. Genetic underpinnings of white matter ‘connectivity’: Heritability, risk, and heterogeneity in schizophrenia. Schizophr Res. 2014 doi: 10.1016/j.schres.2014.03.034. Epub. [DOI] [PubMed] [Google Scholar]
- 28.Kelly S, Morris DW, Mothersill O, Rose EJ, Fahey C, O’Brien C, et al. Genome-wide schizophrenia variant at MIR137 does not impact white matter microstructure in healthy participants. Neurosci Lett. 2014;574:6–10. doi: 10.1016/j.neulet.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Liu B, Zhang X, Hou B, Li J, Qiu C, Qin W, Yu C, Jiang T. The impact of MIR137 on dorsolateral prefrontal-hippocampal functional connectivity in healthy subjects. Neuropsychopharmacology. 2014;39:2153–60. doi: 10.1038/npp.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Erp TG, Guella I, Vawter MP, Turner J, Brown GG, McCarthy G, et al. Schizophrenia miR-137 locus risk genotype is associated with dorsolateral prefrontal cortex hyperactivation. Biol Psychiatry. 2015;75:398–405. doi: 10.1016/j.biopsych.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cummings E, Donohoe G, Hargreaves A, Moore S, Fahey C, Dinan TG, et al. Mood congruent psychotic symptoms and specific cognitive deficits in carriers of the novel schizophrenia risk variant at MIR-137. Neurosci Lett. 2013;532:33–8. doi: 10.1016/j.neulet.2012.08.065. [DOI] [PubMed] [Google Scholar]
- 32.Gur RE, Maany V, Mozley PD, Swanson C, Bilker W, Gur RC. Subcortical MRI volumes in neuroleptic-naïve and treated patients with schizophrenia. Am J Psychiatry. 1998;155:12. doi: 10.1176/ajp.155.12.1711. [DOI] [PubMed] [Google Scholar]
- 33.Brandt GN, Bonelli RM. Structural neuroimaging of the basal ganglia in schizophrenic patients: a review. Wien Med Wochenschr. 2008;158:84–90. doi: 10.1007/s10354-007-0478-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.