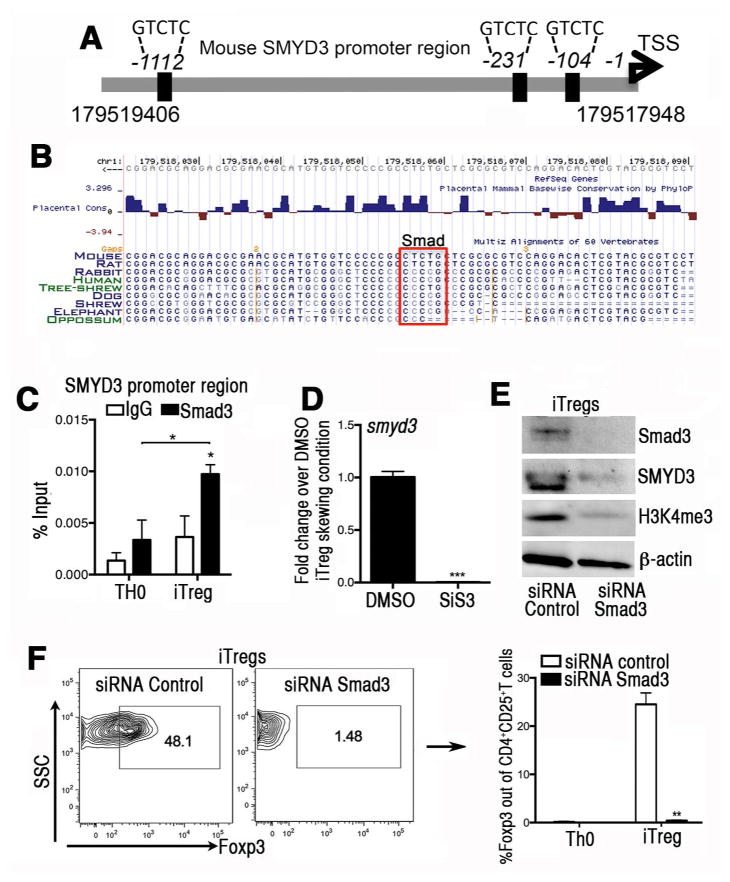
Figure 3. The induction of SMYD3 in iTregs is mediated in a Smad3-dependent manner.
(A) Schematic of the murine SMYD3 promoter region (accession # NM_027188) highlighting potential putative Smad3 binding sites at −104bp, −231 bp, and −1112 bp upstream of the SMYD3 transcription starting site. (B) A Smad3 binding site is evolutionary conserved within the minimal SMYD3 promoter (104bp). Conserved Smad3 binding site on SMYD3 promoter extracted from USCS Genome browser on Mouse (GRCm38/mm10) Assembly. (C) Cells were cultured in complete RPMI under TH0 or iTreg cell skewing conditions for 18 hours. Chromatin immunoprecipitation (ChIP)-quantitative PCR was performed in the SMYD3 promoter site using Smad3 antibody or species-matched IgG as control. Data show four replicates and are representative of three independent experiments (*P<0.05). (D) Naïve CD4+T cells were isolated and stimulated under iTreg differentiation conditions in the presence of the Smad3 inhibitor SiS3 (10 μm) or DMSO for 4 days and assessed for SMYD3. (E) Western blots of Smad3, SMYD3, H3K4me3 and β-actin from Smad3 or control siRNA transfected T cells under iTregs skewing conditions. (F) Foxp3 protein levels in naïve CD4+T cells transfected with Smad3 or control siRNA under iTreg skewing condition. Data are means ±SD and are representative of three independent experiments. *P<0.05, **P<0.01, ***P<0.001.