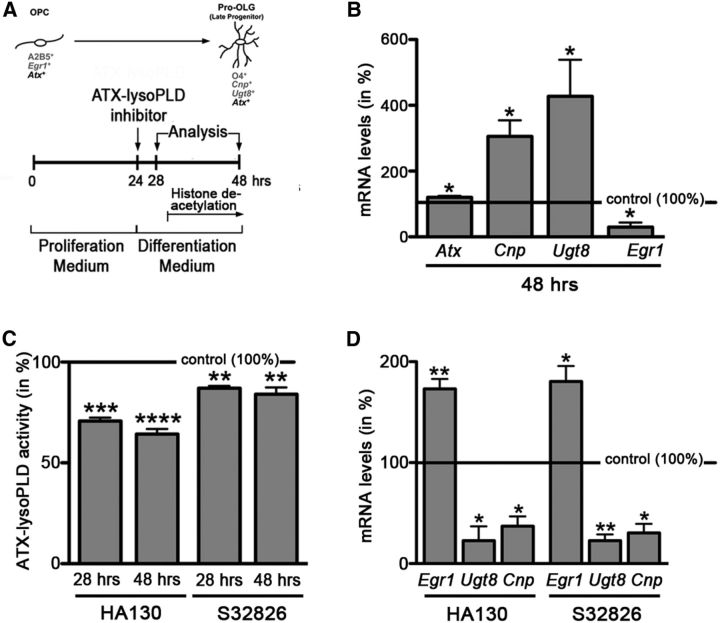
Figure 5.
In rodent OLG cultures, inhibition of the lysoPLD activity of ATX leads to a reduction in the levels of mRNAs encoding OLG differentiation genes and to an increase in the level of mRNA encoding the transcriptional OLG differentiation inhibitor Egr1. A, Experimental design. Timing of histone deacetylation is marked as described by Marin-Husstege et al. (2002). OPC, OLG progenitor. B, Bar graph depicting mRNA levels for the OLG differentiation genes Cnp and Ugt8 and the transcriptional inhibitor Egr1, as well as Atx as determined by real-time RT-qPCR analysis. mRNA levels at 24 h were set to 100% for each gene, and 48 h values were calculated accordingly. C, Bar graph illustrating ATX–lysoPLD activity as assessed by using the fluorogenic substrate FS-3. Control (vehicle-treated) values were set to 100% (see horizontal line), and experimental values were calculated accordingly. D, Bar graph depicting mRNA levels for the OLG differentiation genes Cnp and Ugt8 and the transcriptional inhibitor Egr1 as determined by real-time RT-qPCR analysis. Control (vehicle-treated) values were set to 100% (see horizontal line), and experimental values were calculated accordingly. Data shown in all bar graphs represent means ± SEMs. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.