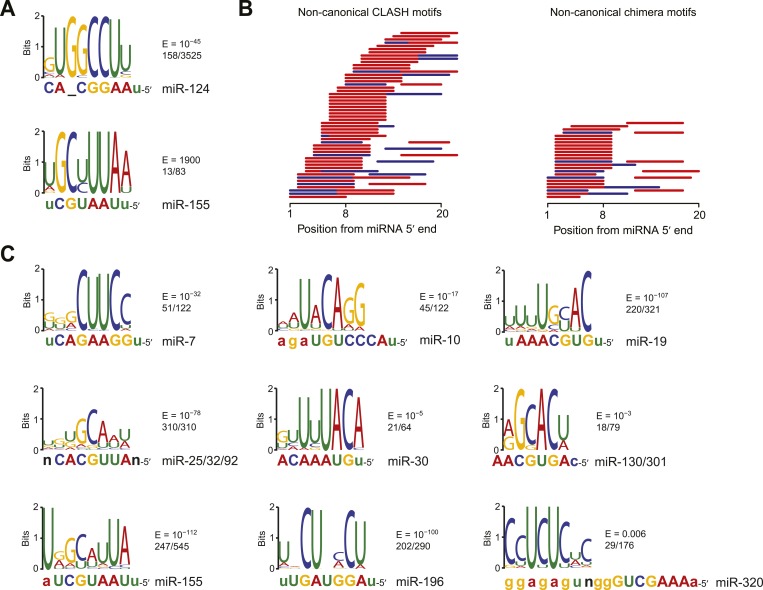
Figure 2. Confirmation of experimentally identified non-canonical miRNA binding sites.
(A) Sequence logos corresponding to motifs enriched in dCLIP clusters that either appear following transfection of miR-124 into HeLa cells (Chi et al., 2009) (top) or disappear following knockout of miR-155 in T cells (Loeb et al., 2012) (bottom). Shown to the right of each logo is its E-value among clusters lacking a seed-matched or offset-6mer canonical site and the fraction of these clusters that matched the logo. Shown below each logo are the complementary regions of the cognate miRNA family, highlighting nucleotides 2–8 in capital letters. (B) Position of the top-ranked motif corresponding to non-canonical sites enriched in CLASH (Helwak et al., 2013) (left) or chimera (Grosswendt et al., 2014) (right) data for each human miRNA family supported by at least 50 interactions without a seed-matched or offset-6mer canonical site. For each family the most enriched logo was aligned to the reverse complement of the miRNA. In cases in which a logo mapped to multiple positions along the miRNA, the positions with the best and second best scores are indicated (red and blue, respectively). (C) Sequence logos of motifs enriched in chimera interactions that lack canonical sites. As in (A), but displaying sequence logos identified in the chimera data of panel (B) for a sample of nine human miRNAs. Logos identified for the other human miRNAs are also provided (Figure 2—figure supplement 1B). A nucleotide that differs between miRNA family members is indicated as a black ‘n’.