Abstract
Macrophages exhibit phenotypic diversity permitting wide-ranging roles in maintaining physiologic homeostasis. Hyaluronic acid, a major glycosaminoglycan of the extracellular matrix, has been shown to have differential signaling based on its molecular weight. With this in mind, the main objective of this study was to elucidate the role of hyaluronic acid molecular weight on macrophage activation and reprogramming. Changes in macrophage activation were assessed by activation state selective marker measurement, specifically quantitative real time polymerase chain reaction, and cytokine enzyme-linked immunoassays, after macrophage treatment with differing molecular weights of hyaluronic acid under four conditions: the resting state, concurrent with classical activation, and following inflammation involving either classically or alternatively activated macrophages. Regardless of initial polarization state, low molecular weight hyaluronic acid induced a classically activated-like state, confirmed by up-regulation of pro-inflammatory genes, including nos2, tnf, il12b, and cd80, and enhanced secretion of nitric oxide and TNF-α. High molecular weight hyaluronic acid promoted an alternatively activated-like state, confirmed by up regulation of pro-resolving gene transcription, including arg1, il10, and mrc1, and enhanced arginase activity. Overall, our observations suggest that macrophages undergo phenotypic changes dependent on molecular weight of hyaluronan that correspond to either (1) pro-inflammatory response for low molecular weight HA or (2) pro-resolving response for high molecular weight HA. These observations bring significant further understanding of the influence of extracellular matrix polymers, hyaluronic acid in particular, on regulating the inflammatory response of macrophages. This knowledge can be used to guide the design of HA-containing biomaterials to better utilize the natural response to HAs.
Keywords: macrophage, hyaluronic acid, molecular weight, classically activated, alternatively activated, polarization
Introduction
Since its discovery,1 hyaluronic acid (HA) has received great attention as a versatile and highly functional biopolymer.2-6 HA is evolutionarily conserved from simple prokaryotes all the way to complex eukaryotes, which is an undeniable testament to its biologic relevance.7 Structurally, hyaluronic acid is not inert. The native, high molecular weight, form can be broken down into smaller molecular weight fragments in response to glycosidase activity upregulated by environmental cues, such as pH and reactive oxygen species.2, 8 The molecular weight variants have been used in a variety of biomedical applications eliciting varying biologic responses.9-10 Clinically, HA has been used in several applications, including ophthalmology as a drug delivery system,11-12 in osteoarthritis for viscosupplementation,13-15 and as a dermal filler.16-17
Physiologic responses to HA are, in part, mediated through the immune system, resulting in either acute or chronic inflammation,18 manifested by the production of specific inflammatory mediators.19-24 Low molecular weight HA promotes the production of inflammatory mediators.25-28 Similarly, high molecular weight HA inhibits production of pro-inflammatory mediators,25, 29-31 suggesting differential macrophage activation by different molecular weight HAs. However, to our knowledge, there has been no work clearly elucidating the role of hyaluronic acid molecular weight in reprogramming of resting, classically activated, and alternatively activated macrophages. An understanding of macrophage reprogramming in response to HA of various molecular weights would establish a clear role for the biomaterial within the context of disease and for the production of biologically responsive biomaterial based systems.32-33
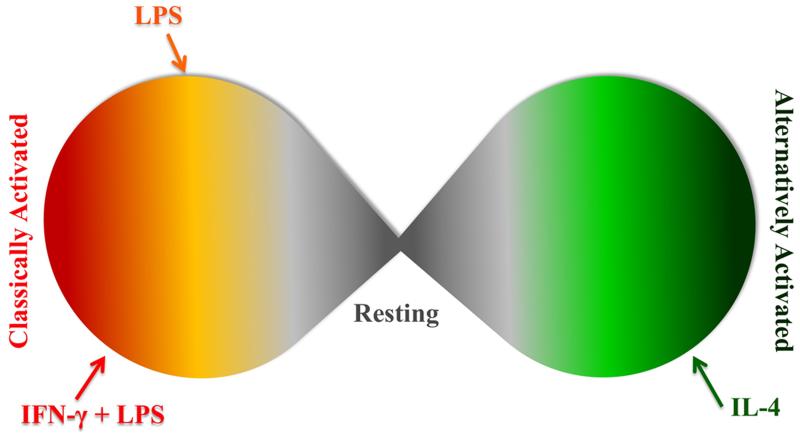
Nominally, there are two extremes of macrophage activation in vitro (Figure 1), which are, in part, mediated by T-helper cell responses to stimuli.33-35 In vivo, the definitions of classically activated macrophages (cMϕs) and alternatively activated macrophages (aMϕs) are understood to be theoretical extremes and the complexity of in vivo signaling results in mixed macrophage populations exhibiting characteristics of both activation states.33 Classically activated macrophages produce and release a robust milieu of inflammatory mediators,32, 34-36 resulting in a destructive environment, evolutionarily designed to purge infecting pathogens or respond to danger signals.
Figure 1. Macrophage activation is a continuum.
Classically and alternatively activated macrophages are two extremes of a continuous spectrum. Many stimuli have the ability to polarize macrophages and even stimuli that give generally same phenotype (LPS or IFN-γ with LPS) do not result in the same activated state.
On the opposing extreme are aMϕs, which are involved in the resolution of inflammation, extracellular matrix reconstruction, and angiogenesis.37-40 The classification as “alternatively” activated has become rather loose, and some use it to refer to any activation state, which is not classical.32, 37 We refer to alternative activation as described when IL-4 was found to induce a novel form of activation in macrophages.37
Although these are two extremes of the macrophage spectrum, the macrophage is multi-faceted in its polarization capability and often exists within a spectrum of these extremes (Figure 1).32-33 Therefore, macrophage activation state can be characterized by the expression of both pro-inflammatory and pro-resolving markers. Herein, we have explored the effect of different hyaluronic acid molecular weights, low (≤ 5 kDa), intermediate (60-800 kDa), and high (> 800 kDa), on macrophage activation. Additionally, we have explored the role of initial macrophage polarization state in mediating response to these various molecular weights to assess reprogramming potential. We have analyzed the expression of inflammatory and pro-resolving markers to unravel and clarify the effects of HA molecular weight on macrophages and compared these results with the initial activation state (Figure S1). We hypothesized that macrophages, either activated or not, would differentially respond to HAs of different molecular weights.
Methods
Hyaluronic acids and hyaluronic acid digestion
Hyaluronic acid, molecular weights 5 kDa, 60 kDa, 800 kDa, and 3,000 kDa, was purchased from Lifecore Biomedical as lyophilized powder (Pharmaceutical Grade, endotoxin ≤ 0.01 EU/mg, Figure S2). Polydispersity (Mw/Mn) of each of these HAs was narrow, not exceeding 1.3 for any of the polymers. Hyaluronic acid digests were formed as previously described with modification.41 Briefly, 50 mg of high molecular weight HA (3,000 kDa) was dissolved in 10 mL of phosphate buffered saline. Mammalian (bovine testicular) hyaluronidase (Sigma-Aldrich, St Louis, MO, USA) was added at a concentration of 10 U/mg of HA, and reacted at 37°C for two hours, after which the temperature was raised to 95°C for twenty minutes. The solution was dialyzed (MWCO 3,500 g/mol) against distilled deionized water for three days. The dialysate was collected and filtered through a 0.22 μm filter, then lyophilized for use. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectroscopy was performed using a Voyager DE PRO Mass Spectrometer (Applied Biosystems, Foster City, CA) equipped with a 337-nm pulsed (~ 5 nsec) nitrogen laser42-43 revealed a polydisperse sample with average molecular weight of approximately 1.3 kDa (not shown), and molar concentration of the digests was determined using this average molecular weight. The limulus amebocyte lysate (LAL; Pierce) assay was used to confirm the absence of endotoxin in the digest preparation according to manufacturer’s instructions.
A HA concentration of 1 μM was chosen for all experiments since therapeutic pharmaceutical formulations are administered at approximately this concentration.44-46 Similarly, most investigative biomaterial based systems utilize similar concentrations, and many organs with high infiltration of macrophages during injury, such as extracellular matrix,30 vitreous body,47-48 pleural cavity,49 lymphatic system,30 and joint space50, contain similar concentrations of HA.
Cell culture and treatments
Murine macrophages (J774A.1 ATCC; TIB-67)51-52 were cultured in phenol-free Dulbecco’s modified Eagle’s medium (DMEM; Hyclone, Logan, UT, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; Gemini, Calabasas, CA, USA) at 37°C in 5% CO2. Macrophages were harvested by scraping and used between passage 2 and 7. Macrophages were plated at a density of 5×105 cells/mL in a 24 well plate (500 μL/well) and allowed to adhere overnight before use.
Four states of macrophages were used in each experiment. Macrophages were cultured in the presence or absence of varying molecular weights (5-3,000 kDa, or the digest) of hyaluronic acid (1μM) (1) in their resting state, i.e. cultured in media with no activating agent (M), (2) with 100 ng/mL γ-irradiated LPS (M(LPS)) derived from Escherichia coli serotype 055:B5 (Sigma-Aldrich, St Louis, MO, USA), (3) with 20 ng/mL interferon-γ (Peprotech) and 100 ng/mL γ-irradiated LPS (M(IFN-γ+LPS)),36 or (4) following activation with 20 ng/mL interleukin-4 (Peprotech) (M(IL-4))37. In each experiment scenario, the reference state—M, M(LPS), M(IFN-γ+LPS), or M(IL-4)—macrophages were cultured for the same time as the experimental groups without HAs added (Figure S1).
Nitric oxide (NO)
J774A.1 murine macrophages were plated with treatments as described above. Supernatant media was collected after 24-hour incubation period and nitrite measured using the Griess reagent (Promega, Madison, WI, USA) according to manufacturer’s instructions.
Arginase activity
Arginase activity was assayed as previously described53-54 Briefly, lysates of 105 macrophages cultured as described above were centrifuged at 12,000×g for 20 minutes and the supernatant was collected. Arginase in the supernatant was activated by heating the lysate for 10 minutes at 55°C in a 50 mM Tris-HCl buffer (pH 7.5) containing 10 mM MnCl2. L-arginine hydrolysis was then carried out with the addition of 25 μL of 0.5 M L-arginine (pH 9.7) to 25 μL of the activated lysate and incubated at 37°C for 60 minutes. The reaction was stopped with the addition of an acid mixture containing H2SO4, H3PO4, and H2O (1:3:7). After the addition of 25 μL of a 9% solution of α-isonitrosopropiophenone and heating to 100°C for 45 minutes, the end product of the hydrolysis reaction, urea was detected colorimetrically on a Labsystems Multiskan plus spectrophotometric microplate reader (Thermo Scientific, Hanover Park, IL) at 540 nm.
TNF-α production
Tumor necrosis factor- α (TNF-α) secreted by macrophages was determined by the measuring the protein in cell-culture supernatants using a TNF-α enzyme-linked immunosorbent assay kit (R&D systems) according to the manufacturer’s instructions.
Gene expression
TRIzol (Invitrogen, Carlsbad, CA, USA) was used to isolate total RNA from J774A.1 murine macrophages. Integrity of RNA was determined via spectrophotometric analysis of each sample at 260 and 280 nm. Samples with 260 to 280 ratios greater than 1.8 were reverse transcribed into cDNA (Applied Biosystems, Carlsbad, CA, USA). Real time PCR was carried out on an Applied Biosystems StepOnePlus™ PCR using SYBR® Green PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA). A melting curve analysis was performed after each run to confirm product specificity. All primers were designed to span exon-exon junctions (Table 1). Transcripts of β-glucuronidase (gusb) were quantified and used as endogenous control.55 Relative quantities were estimated by the delta-delta-Ct method.56 The expression of each gene was normalized to untreated cells as control.
Table 1.
Genes and primers used for quantitative real-time PCR.
| Gene | Protein | Acronym | Primers (5′→3′) | Accession number |
|---|---|---|---|---|
| gusb | glucuronidase, beta | GUSB | FW: GCAAGACATCGGGCTGGTGA REV: TGGCACTGGGAACCTGAAGT |
NM_010368.1 |
| nos2 | nitric oxide synthase 2, inducible |
iNOS | FW: AGCCCCGCTACTACTCCATC REV: GCCACTGACACTTCGCACAA |
NM_010927.3 |
| tnf | tumor necrosis factor | TNF-α | FW: AACTTCGGGGTGATCGGTCC REV: TGGTTTGTGAGTGTGAGGGTCT |
NM_001278601.1 |
| il12b | interleukin 12b | IL-12 | FW: GAACTGGCGTTGGAAGCACG REV: GGCGGGTCTGGTTTGATGAT |
NM_008352.2 |
| il10 | interleukin 10 | IL-10 | FW: CTCCTAGAGCTGCGGACTGC REV: GGCAACCCAAGTAACCCTTAAAGT |
NM_010548.2 |
| arg1 | arginase | Arg1 | FW: AAGACAGGGCTCCTTTCAGGAC REV: TCCCGTTGAGTTCCGAAGCA |
NM_007482.3 |
| cd80 | cluster of differentiation-80 |
CD80 | FW: GAAAAACCCCCAGAAGACCCTC REV: TGACAACGATGACGACGACTGT |
NM_009855.2 |
| mrc1 | mannose receptor, C type 1 |
Mrc1 | FW: CAGACAGGAGGACTGCGTGG REV: TGCCGTTTCCAGCCTTTCCG |
NM_008625.2 |
Statistical Analyses
ANOVA was used to test all groups, and post-hoc Tukey analysis was utilized if ANOVA suggested significant differences between the groups with significant p-values less than 0.05. Statistical significant differences from the reference states (Figure S2) are presented with the average value for that reference state presented in the figures with a dotted line of appropriate color for that state (resting, LPS, LPS with IFN-γ, or IL-4). All experiments were independently replicated at least three times.
Results and Discussion
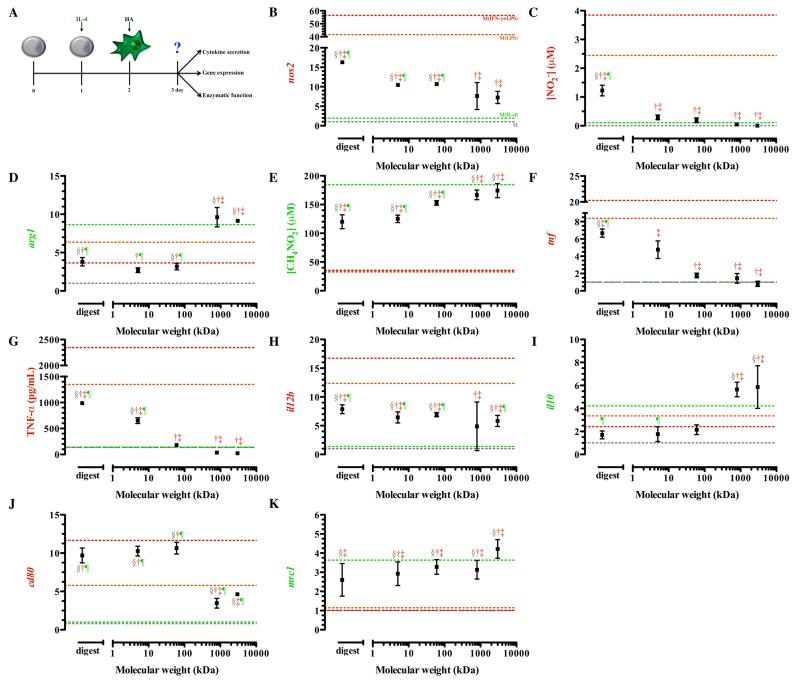
Resting Macrophage: Stimulation with Hyaluronic Acid
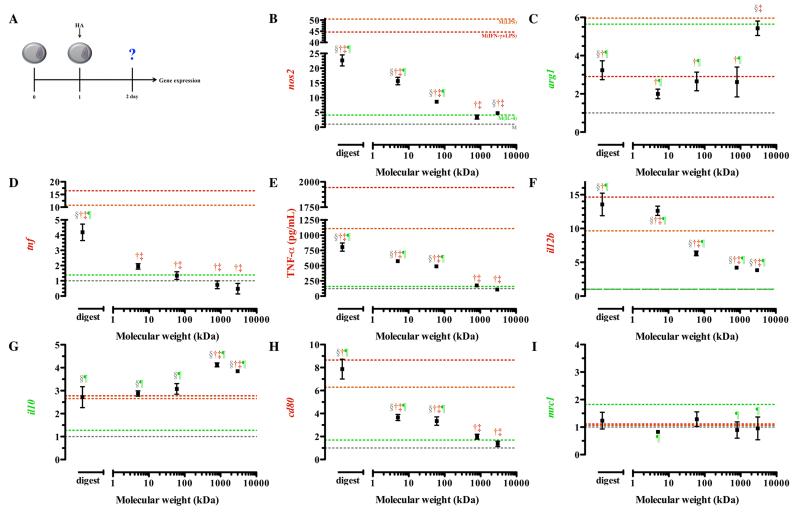
Our initial assessment of macrophage phenotype in response to hyaluronic acid was done on resting macrophages (Figure 2A). We observed that the transcription of nos2 was significantly up-regulated upon treatment with all molecular weights of HA, except 800 kDa HA compared to resting macrophages (Figure 2B). Lower molecular weight HAs induced the greatest increase in nos2 expression, with the digestion showing an over 20-fold increase from resting state (Figure 2B). Although lower molecular weights increased nos2, this was significantly less than M(LPS) and M(IFN-γ+LPS). Higher molecular weight HAs (800 and 3,000 kDa) exhibited nos2 expression levels comparable to M(IL-4) (p > 0.05).
Figure 2. Gene expression of resting macrophages in response to hyaluronic acid.
Resting macrophages were treated with 1μM HAs for 24 hours before phenotypic assessment was made. The schematic depicts the time course of the experiments (A). Low molecular weight HAs induced transcription of nos2 (B), decreased arg1 transcription (C), and enhanced tnf transcription (D) and TNF-a (E). High molecular weight HAs stimulated minimal il12b (F) transcription and enhanced il10 (G) transcription. Low molecular weight HAs enhanced transcription of cd80 (H) but did not change mrc1 (I) transcription in resting macrophages. Points represent mean plus or minus (±) standard deviation of three independent experiments. The symbols indicate significant difference (p-value less than 0.05) from the reference states, which are represented by corresponding colored dotted lines: resting macrophages (§; M), classically activated macrophages, stimulated with LPS (†; M(LPS)) or LPS and IFN-γ (‡; M(IFN-γ+LPS)), or alternative activated macrophages, stimulated with IL-4 (¶; M(IL-4)).
Arginase expression was significantly up-regulated in macrophages treated with 3,000 kDa HA compared to resting macrophages (p < 0.0001) (Figure 2C). This was not significantly different from expression detected in M(IL-4) and is suggestive of movement to an alternatively activated state. Differences in functional by-products for both of these enzymes were undetectable after treatment for 24 hours (data not shown).
Upon treatment with the digest, macrophages exhibited a 4-fold increase in the amount of tnf gene transcription compared to resting macrophages (Figure 2D). All other molecular weights tested were not statistically different from resting macrophages, indicating that only oligomeric HA modulates transcription of tnf (Figure 2D). TNF-α production was minimal in macrophages treated with the highest molecular weight HAs (800 and 3,000 kDa), and the secreted levels were comparable to M(IL-4) (p > 0.05) (Figure 2E).
Next, we examined the expression of il12b and il10. Macrophages treated with all HA molecular weights exhibited significantly higher transcription of il12b compared to both resting macrophages and M(IL-4)(Figure 2F). Resting macrophages treated with lower molecular weight HAs (5kDa and the digest) expressed il12b that exceeded that of M(LPS). Macrophages treated with intermediate and high molecular weight HAs transcribed lower levels of il12b compared to M(LPS) and M(IFN-γ+LPS). Transcription of il10 was significantly up-regulated in resting macrophages treated with all HAs compared to their native state. Macrophages treated with the lowest molecular weight HAs (digest, 5, and 60 kDa) showed no significant difference in il10 expression compared to M(LPS) and M(IFN-γ+LPS) (Figure 2G), suggesting polarization that is similar to the classically activated state. HAs with molecular weight of 800 kDa or 3,000 kDa, though significantly deviating from M(IL-4), had highly enhanced transcription of il10 (Figure 2G), suggesting a movement toward the alternatively activated state.
Finally, we examined the transcription of two phenotype specific surface markers: cd80 and mrc1. High molecular weight HAs (800 and 3,000 kDa) reduced expression of cd80 to levels comparable to M(IL-4) (Figure 2H). On the other hand, the digestion of HA enhanced the expression of cd80 to levels associated with the classically activated state (p > 0.05; Figure S1J, 2H). Transcription of mrc1 was minimally influenced by HA treatment (Figure 2I). Overall, it was clear that resting macrophage exposure to lower molecular weight HAs resulted in activation similar to cMϕs while higher molecular weight HAs stimulated activation similar to aMϕs.
Classically Activated Macrophages: Stimulation with LPS and HA simultaneously
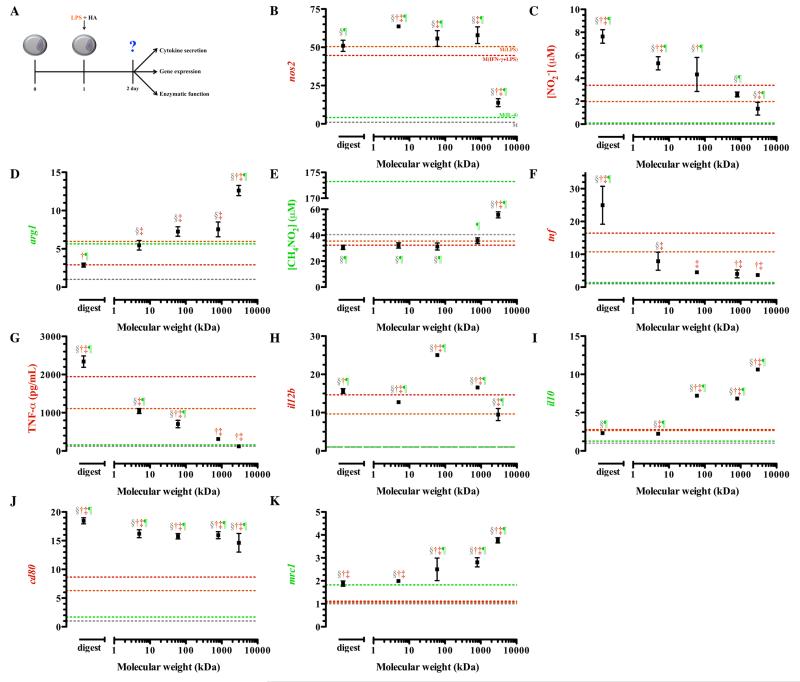
In an effort to mimic an inflammatory response that may occur simultaneous to administration of HA, we examined simultaneous administration of HA and LPS (Figure 3A).57 Delivery of lipopolysaccharide derived from E. coli cell wall was chosen to simulate this response in vitro.58 LPS activates macrophages through a pathway, which is shared with other biomaterials, in vitro as well as in vivo, so we considered this an adequate in vitro method of simulating an acute classical inflammatory response.57-61 Previous studies have established that high molecular weight hyaluronic acid has the ability to suppress inflammation due to LPS.62 However, molecular weight dependent effects on specific phenotype markers have not been clarified.
Figure 3. Gene expression, cytokine secretion, and enzymatic function of classically activated macrophages (M(LPS) treated simultaneously with HAs.
After plating, macrophages were simultaneously treated with LPS and HAs. The schematic depicts the time course of the experiments (A). Expression of nos2 (B) and nitrite production (C) was elevated in groups treated with low molecular weight HAs. Treatment of macrophages with LPS and high molecular weight HAs enhanced arg1 (D) transcription and production of urea (E). Expression (F) and secretion (G) of tumor necrosis factor alpha was lowest in groups treated with high molecular weight HAs. Expression of il12b (H) and cd80 (J) was elevated in groups treated with low molecular weight HAs. Transcription of il10 (I) and mrc1 (K) steadily increased with increasing molecular weight. Points represent mean plus or minus (±) standard deviation of at least three independent experiments. The symbols indicate significant difference (p-value less than 0.05) from the reference states, which are represented by corresponding colored dotted lines: resting macrophages (§; M), classically activated macrophages, stimulated with LPS (†; M(LPS)) or LPS and IFN-γ (‡; M(IFN-γ+LPS)), or alternative activated macrophages, stimulated with IL-4 (¶; M(IL-4)).
Transcription of nos2 was significantly increased upon treatment with all HA molecular weights except for 3,000 kDa HA (Figure 3B). This increase matched or exceeded transcription levels of nos2 for M(LPS) and M(IFN-γ+LPS). Macrophages treated with the highest molecular weight HA (3,000 kDa) decreased nos2 transcription to levels approaching levels of M(IL-4). Low (digest and 5 kDa) and intermediate molecular weight HAs (60 kDa) significantly enhanced the production of nitrite compared to untreated LPS stimulated macrophages (Figure 3C) while arg1 expression steadily increased with increasing molecular weight HA.
The highest levels of arg1 transcription were detected in macrophages treated with 3,000 kDa HA, and these expression levels significantly exceeded the levels of M(IL-4) (Figure 3D). Treatment with low and intermediate HAs resulted in arg1 levels that were not different from M(LPS) (Figure 3D). In macrophages treated with HA with molecular weight less than 3,000 kDa, arginase activity was comparable to M(LPS) (Figure 3E). These HAs also suppressed arginase activity to the same extent as found in both M(LPS) and M(IFN-γ+LPS). Macrophages treated with the highest molecular weight HA, 3,000 kDa, resulted in significantly elevated urea production compared to macrophages treated with LPS (Figure 3E). However, this level of urea production was still significantly lower than M(IL-4) (Figure S1E).
Only the lowest molecular weight (digests) and the highest molecular weight HA (3,000 kDa) showed statistically different tnf transcription compared to M(LPS) (Figure 3F). The HA digest promoted expression of tnf, inducing a robust 24-fold increase in tnf expression compared to untreated cells and a 14-fold increase in expression compared to M(LPS) (Figure 3F). On the other hand, treatment with high molecular weight HA (3,000 kDa) decreased the expression tnf from M(LPS) by about 7-fold (Figure 3F). TNF-α protein secretion exhibited a similar trend to tnf gene expression. Macrophages treated with the HA digest exhibited TNF-α release that was 2.5 times greater than M(LPS) (Figure 3G). On the other hand, macrophages treated with 3,000 kDa HA decreased TNF-α secretion to approximately half that secreted by M(LPS).
Macrophages treated with all molecular weight HAs, except 3,000 kDa, resulted in significantly enhanced expression of il12b compared to M(LPS) (Figure 3H). Transcription of il12b did not suggest differential polarization in response to HAs; however, differences in pro-resolving il10 were much more defined by molecular weight, with increasing molecular weight HAs enhancing the expression of this pro-resolving gene (Figure 3I). The greatest expression of il10 was observed following treatment with 3,000 kDa, suggesting that higher molecular weight HAs enhance pro-resolving function and activate macrophages similar to M(IL-4).
To further confirm this trend, we analyzed surface markers mRNA expression for cd80 and mrc1 in this population of macrophages. All HA molecular weights significantly increased cd80 transcription compared to M(LPS) and M(IFN-γ+LPS) (Figure 3J). However, mrc1 expression, a marker of alternative activation, was also up-regulated in macrophages exposed to higher molecular weight HAs (Figure 3K) suggesting that these molecular weight HAs enhance expression of both classical- and alternative-associated surface receptors. Moreover, lower molecular weight HAs (digest and 5kDa) expressed each surface marker to and extent that matched or exceeded that of M(LPS) and M(IFN-γ+LPS).
Classically Activated Macrophage: Stimulation with HAs
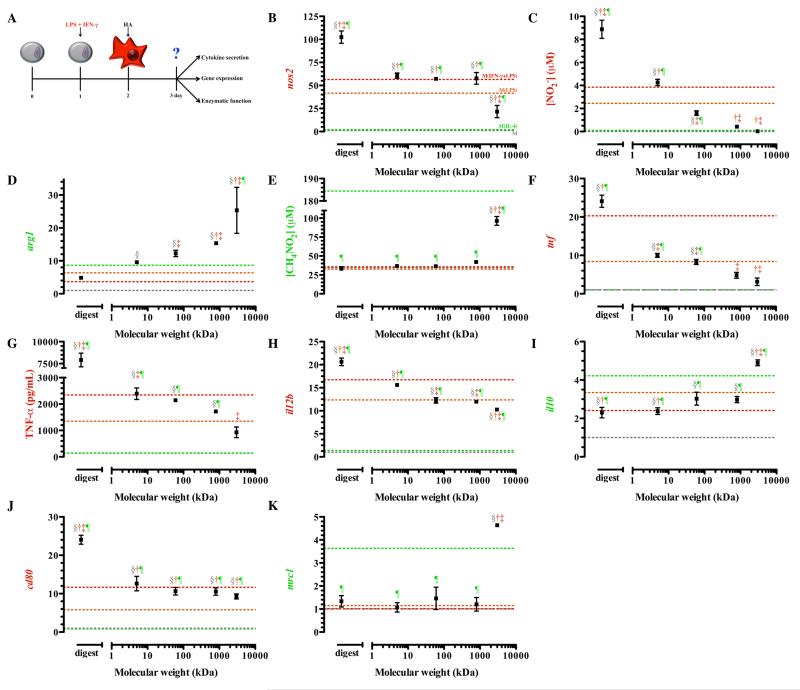
Chronic diseases are often associated with persistently activated macrophages, whether in the alternative or classically activated state32, 34-36, 39-40 but, cMϕs are considered to have the most detrimental effects. Therefore, we explored the modification macrophage phenotype with HA in a cMϕ population, produced in the presence of both IFN-γ and LPS to represent a more sustained and robust inflammatory response.63 Modification of such a population would not only suggest therapeutic potential in reprogramming macrophage phenotype, but also, a potential role for HA in the propagation or alteration of macrophage function within the tissue.
Upon treatment with the HA digest, M(LPS) and M(IFN-γ+LPS) exhibited significantly enhanced transcription of nos2. Classically activated macrophages treated with 3,000 kDa HA attenuated nos2 transcription, although this remained significantly higher than M(IL-4) (Figure 4B). Classically activated macrophages treated with the lowest molecular weight HAs, the digests and 5 kDa, produced significantly more nitrite than M(IFN-γ+LPS) not treated with HAs (Figure 4C). On the other hand, cMϕs treated with 800 to 3,000 kDa HA suppressed production of nitrite to levels measured in M(IL-4) (Figure 4C). Biologically significant decreases in nitric oxide produced by M(IFN-γ+LPS) were dependent on treatment of HA with molecular weight greater than 60 kDa, with higher molecular weight HAs having the greatest reduction in nitrite production.
Figure 4. Gene expression, cytokine secretion, and enzymatic function of classically activated macrophages (M(IFN-γ+LPS )) treated with HAs.
After plating, macrophages were classically activated with interferon-γ and LPS for 24 hours, after which media was replaced with media containing HAs for treatment (A). Expression of nos2 (B) and production of nitrite (C) was elevated in groups treated with low molecular weight HAs, whereas arg1 (D) and urea production (E) were elevated in groups treated with high molecular weight HAs. (B). Expression (F) and secretion (G) of tumor necrosis factor alpha was significantly reduced in groups treated with high molecular weight HAs. Expression of il12b (H) was elevated in groups treated with lower molecular weight HAs. High molecular weight HAs potentiated il10 expression (I), but did not change cd80 (J) or mrc1 (K) expression. Points represent mean plus or minus (±) standard deviation of at least three independent experiments. The symbols indicate significant difference (p-value less than 0.05) from the reference states, which are represented by corresponding colored dotted lines: resting macrophages (§; M), classically activated macrophages, stimulated with LPS (†; M(LPS)) or LPS and IFN-γ (‡; M(IFN-γ+LPS)), or alternative activated macrophages, stimulated with IL-4 (¶; M(IL-4)).
Arginase was significantly up-regulated to extents similar to M(IL-4) in macrophages treated with all molecular weight HAs. The highest increase in arg1 transcription was detected in macrophages treated with HA 3,000 kDa (Figure 4D), which was significantly higher than M(IL-4). The ability of HA 3,000 kDa to significantly reduce nos2 expression and enhance arg1 expression was consistent with an alternatively activated phenotype. Urea production was significantly elevated, by approximately 3 fold, in macrophages treated with 3,000 kDa HA compared to untreated M(IFN-γ+LPS) (Figure 4E) while M(IFN-γ+LPS)treated with the digest and 5, 60 and 800 kDa HAs produced urea to the same extent as M(IFN-γ+LPS) not treated with the HAs (Figure 4E).
TNF-α expression and production demonstrated that treatment of the macrophages with HA molecular weights less than 5 kDa resulted in augmented transcription of tnf, 10 to 25-fold over untreated macrophages (Figure 4F). Classically activated macrophages treated with 5 through 3,000 kDa HAs significantly reduced the expression of tnf (Figure 4F) compared to the untreated M(IFN-γ+LPS); however, only 800 and 3,000 kDa HAs diminished transcription to levels observed with the M(IL-4) (Figure S1F). Production of TNF- α followed similar pattern to mRNA transcription. Classically activated macrophages treated with the digestion of HA produce up to 3.5 times more TNF-α compared to untreated M(IFN-γ+LPS) (Figure 4G). Higher molecular weight HA, 3,000 kDa, suppressed production of TNF-α to levels observed for M(IL-4).
Transcription of il12b was significantly reduced in M(IFN-γ+LPS) treated with higher molecular weight HAs, with the greatest reduction, approximately 5-fold decrease from the level of cMϕs upon treatment with 3,000 kDa HA (Figure 4H). Unlike il12b transcription, transcription of il10 mRNA was elevated with increasing molecular weight (Figure 4I). The highest molecular weight HA, 3,000 kDa, promoted il10 transcription, which significantly exceeded that of M(IL-4), once again suggesting that treatment with high molecular weight HA resulted in activation similar to aMϕs (Figure 4I).
Classically activated macrophages treated with HA digests enhanced cd80 transcription to levels significantly greater than M(IFN-γ+LPS) (Figure 4J). All other molecular weight HAs did not result in activation that significantly deviated from the cd80 levels of M(IFN-γ+LPS) (Figure 4J). Transcription of mrc1 remained at a basal level in M(IFN-γ+LPS) (Figure 4K) and was only significantly elevated after treatment with HA 3,000 kDa and not significantly different from levels of M(IL-4) (p > 0.05).
Alternatively Activated Macrophages: Stimulation with HA
Although cMϕs are considered destructive in many diseases, aMϕs are also associated with several chronic diseases.39-40 Therefore, we examined the effect of HA molecular weight on macrophages of this population. Macrophages were alternatively activated with overnight treatment with IL-4.37
When M(IL-4) were treated with HAs, molecular weights lower than 800 kDa significantly increased transcription of nos2 compared to the untreated M(IL-4) (Figure 5B). The digest and 5 kDa HAs promoted a 10 to 16 fold increase in nos2 transcription compared to M(IL-4). All HA treatments resulted in significantly lower nos2 transcription compared to M(LPS) and M(IFN-γ+LPS) (p < 0.05; Figure 5B). Although nos2 gene expression was significantly increased from baseline levels of M(IL-4) for all macrophage treatments, only the smallest of the HA fragments, the digest, was able to elicit significantly increased nitrite production (p < 0.0001) compared to M(IL-4) (Figure 5C).
Figure 5. Gene expression, cytokine secretion, and enzymatic function of alternatively activated macrophages (M(IL-4)) treated with HAs.
After plating, macrophages were alternatively activated with IL-4 for 24 hours, after which they were washed and media was replaced with media containing HAs for treatment (A). Expression of nos2 was elevated in all treatment groups (B), but nitrite was only elevated in groups treated with the HA digest (C). Transcription of arg1 was enhanced by high molecular weight HAs (D). Treatment with high molecular weight HAs maintained production of urea (E). Expression (F) and secretion (G) of tumor necrosis factor alpha was significantly elevated in groups treated with low molecular weight HAs. Elevated transcription of il12b (H), reduced transcription of il10 (I), enhanced transcription of cd80 (J), and diminished mrc1 transcription (K) was detected in groups treated with low molecular weight HAs. Points represent mean plus or minus (±) standard deviation of at least three independent experiments. The symbols indicate significant difference (p-value less than 0.05) from the reference states, which are represented by corresponding colored dotted lines: resting macrophages (§; M), classically activated macrophages, stimulated with LPS (†; M(LPS)) or LPS and IFN-γ (‡; M(IFN-γ+LPS)), or alternative activated macrophages, stimulated with IL-4 (¶; M(IL-4)).
Arginase (arg1) expression and activity (urea production) was significantly reduced following lower molecular weight HA, 60 kDa or lower, exposure (Figure 5D-E). Higher HA molecular weights (800 and 3,000 kDa) maintained arg1 transcription at levels at untreated M(IL-4) levels. Additionally, urea production by macrophages treated with higher HA molecular weights was no different from M(IL-4), whereas lower molecular weights HAs significantly reduced urea production (Figure 5E). This suggests that 800 kDa and 3,000 kDa HAs may maintain alternative activation, but that lower molecular weights drive macrophage polarization away from the alternative state.
Classically activated macrophages exhibited tnf transcription and production that was amplified upon treatment with low molecular weight HAs. This observation holds true for M(IL-4). Alternatively activated macrophages treated with low molecular weight HAs exhibited 5 to 6 fold increased tnf transcription (Figure 5F) compared to M(IL-4) not treated with HA. TNF-α production was similar to baseline levels in M(IL-4) treated with 60 to 3,000 kDa HAs. Only the lowest molecular weight HAs (digest and 5 kDa) elicited a significantly enhanced secretion of TNF-α compared to untreated M(IL-4) (Figure 5G).
Alternatively activated macrophages treated with 800 kDa HAs did not exhibit changes in il12b transcription compared to untreated M(IL-4) (p > 0.05; Figure 5H). All other molecular weight HAs significantly enhanced il12b transcription. On the other hand, il10 expression was significantly reduced to levels similar to M(LPS) and M(IFN-γ+LPS) in M(IL-4) treated with HAs 60 kDa and lower (Figure 5I). Macrophages treated with 800 to 3,000 kDa HAs showed no differences in il10 from the alternatively activated state.
Alternatively activated macrophages treated with all HAs resulted in elevated cd80 transcription compared to untreated M(IL-4). Low and intermediate molecular weight HAs (less than or equal to 60 kDa) up-regulated cd80 by approximately 10 fold (Figure 5J). Conversely, treatment with higher molecular weight HAs resulted in significantly lower enhancement of cd80 transcription, approximately 5-fold higher compared to M(IL-4) (Figure 5J). Levels of alternative activation marker, mrc1, were maintained nearly constant upon treatment with all HAs and did not significantly deviate from levels associated with M(IL-4) (Figure 5K).
Again, lower molecular weight HAs drove activation away from aMϕs, and toward cMϕs, while higher molecular weight HAs drove activation toward aMϕs, and away from cMϕs. In addition, the relative expression of classical and alternative activation markers (Figure S4-S6) further confirmed the cMϕ and aMϕ phenotypes for all cases, but it is does not appear possible to differentiate resting macrophages from aMϕs using the relative expression of cMϕ and aMϕ markers.
Overall observations
The work presented in this study demonstrates that HA has molecular weight dependent effects on macrophage gene expression, enzyme activity, and cytokine production under a variety of conditions. Based on the interpretation and analysis of several key genes and their products, hyaluronic acid has the ability to modulate and reprogram macrophage phenotype within the wide spectrum of activation states.
We have specifically chosen to look at several key genes and products involved in regulating macrophage response and shown to shape macrophage phenotype. Arginase, urea, il10, and mrc1 are associated with the aMϕ.37-40 Conversely, nos2, nitric oxide, TNF-α, il12b, and cd80 are associated with the cMϕ.36, 64-65 Although the two phenotype paradigm is a simple framework to understand macrophage function, in truth, macrophages reside within a wide spectrum of these activation states and are likely to express all of these genes and markers at once and may express both aMϕ and cMϕ markers simultaneously and their relative expression of several markers (Figures S4-S7) yields information that describes their current position within this complex spectrum of function.32-35
Most macrophage reprogramming approaches have focused on the aMϕ as a desired response, since these macrophages have the ability to decrease inflammation, induce vascularization, promote matrix deposition, and support constructive tissue remodeling.38-39 On the other hand, the cMϕ is often thought of as a mediator of immunopathology and damage. However, the cMϕ is actually very important in host defense against pathogens. Proper clearance of pathogen cannot be done without this activation and is hindered in other activation states.35, 39-40 Classically activated macrophages also secrete tumoricidal cytokines, and spacio-temporal control of this activation may be desirable in the tumor environment.35 Though both macrophage responses are desired in certain situations, it should be noted that abnormal polarization of the macrophage in either direction leads to detrimental function. Our findings suggest that endogenous HA or therapeutically applied HA, i.e. as a biomaterial or drug delivery system, can alter macrophage polarization in a molecular weight-dependent manner.
Our original hypothesis was that macrophage reprogramming depends, in part, upon the initial polarization state. We chose to examine the response of macrophages to HA in four initial polarization states: the resting state and models of classical and alternative activation. In each of these cases, hyaluronic acid has the ability to modulate response dependent on its molecular weight. Based on our results in all four initial polarization states, we can conclude that HA with molecular weights in the megadalton range tend to decrease the expression of markers normally associated with the classical state and enhance or sustain those associated with an alternative activation state. Though the magnitude of response may be different in each case, modulation of the mediators by hyaluronic acid follows a similar trend independent of initial macrophage activation state. This suggests that treatment with these molecular weights may have true reprogramming potential, promoting an alternatively activated-like state. Our results are in concordance with findings that show that administration of exogenous high molecular weight HA promotes resolution phase of wound healing and that removal of this polymer is detrimental to the process.66-68 Our findings also suggest that administration of high molecular weight hyaluronic acid may be therapeutically used to control the phenotype and subsequent activity of macrophages, which are bound in an inflammatory state.
Similarly, we can conclude that HA oligomers smaller than 12 disaccharides in length likely promote a classically activated-like state since they sustain or enhance expression of pro-inflammatory genes and cytokines and reduce those associated with the resolution of inflammation, i.e. the alternative activation state. This too, is independent of initial macrophage activation state. Interestingly, under conditions of classical activation, macrophage treatment with low molecular weight HAs actually augments classically activated markers. Once inflamed, either via classically activating cytokines or other activators, macrophages produce reactive oxygen species and increase the expression of hyaluronidase.8, 27 Both of these are involved in the depolymerization of HAs.8 We observed that cleaved HA products have the highest pro-inflammatory activity. Therefore, treatment of macrophages with HAs under inflammatory conditions may actually promote cleavage of the polymer into even smaller chains, inducing an even greater inflammatory response, thus propagating the classically activated state. Cleavage fo HAs was not examined in this work, and the investigation of the effect of hyaluronidase should be examined in the future. Regardless of the potential for cleavage under inflammatory stress, we observed that treatment with higher molecular weight HAs induced up-regulation of alternatively activated markers, whereas low molecular weight HAs induced the up-regulation of classically activated markers.
Overall, a trend for most genes, enzymes, and cytokines shows that macrophages treated with higher molecular weights exhibit reduced expression of classically activated genes and mediators and augmented expression of alternatively activated markers. The opposite is true for macrophages treated with low molecular weight HAs. In these cases, classical markers are enhanced. The macrophage response to intermediate molecular weights is much more difficult to decipher. Macrophage response upon treatment with these molecular weights is very much dependent on their initial state, suggesting that response to these molecular weights results in (1) a single population of macrophages which simultaneously express both classically activated as well as alternatively activated markers, (2) mixed populations of activated macrophages, or (3) these molecular weights do not have preference for binding to specific receptors that have distinct signaling toward a particular activation state. The intermediate molecular weights may have similar affinities to several receptors, some of which promote classical activation and others that promote alternative activation concomitantly.
Although HA is notoriously promiscuous in its binding to several different receptors and associated proteins, signaling through one of or a combination of mechanisms may be involved in the macrophage reprogramming we observe. The cluster determinant 44 (CD44) receptor is the best-characterized receptor of HA. Stimulation of CD44 with HA has been shown to play a role in cell adhesion, cell-substrate interactions, metastasis, and inflammation.69-71 Pro-inflammatory stimuli, such as IFN-γ, LPS, and TNF-α, induce a high-affinity HA binding state of CD44 by increasing expression of the receptor and inducing certain post-translational modifications, such as reducing chondroitin sulfation and N-glycosylation of CD44.72-75 Priming of macrophages with IFN-γ has been shown to enhance CD44 signaling.76 In contrast, IL-4 induces post-translational modifications of CD44 receptor, which inhibit HA binding.72, 74-75 This observation may explain why classically activated macrophages exhibit augmented response to HAs compared to those alternatively activated. On the other hand, binding of high molecular weight HA to CD44 may be inducing its suppressive and alternatively activating effects through up-regulation of transcription factors.77-79 Additionally, multi-ligand CD44 occupancy, i.e. from high molecular weight hyaluronan, enhances macrophage phagocytosis and clearance of apoptotic cells, a function associated with alternatively activated, pro-resolving, macrophages.80-81 Even with signaling through this one receptor, there are multiple potential mechanisms that need to be examined in addition to other HA receptors that must be considered.
HA fragments have also been shown to signal through toll-like receptor 4 (TLR-4).28, 82-83 This receptor is essential in recognizing pathogen associated molecular patterns, such as motifs in exterior carbohydrates, including as mannose or LPS84 leading to subsequent up-regulation of the expression of several pro-inflammatory genes associated with cMϕs.84 Low molecular weight HAs may be signaling through toll-like receptors to produce the cytokines and up-regulate markers associated with classical activation. It does not appear that high molecular weight HAs signal through TLR-4, and the mechanism of discriminating between low and high molecular weight HA is not known.
Macrophages activated with multiple toll-like receptor ligands tend to show either a sustained or additive classical activation state.85 Our observations fall in line with these discoveries. In our models, macrophages treated simultaneously with low molecular weight HAs and TLR ligands (LPS) exhibit a sustained and sometimes augmented expression and production of inflammatory genes and mediators, which may further suggest that low molecular weight HAs are inducing inflammatory response through toll-like receptors. In contrast to the activating potential of low molecular weight HAs, high molecular weight HAs have been shown to reduce TLR-4 expression in LPS stimulated cells, which may account for its anti-inflammatory activities, as well as its ability to modify macrophage response to a more alternatively activated-like state.86
Other receptors that may be involved in the differential response to HA include the receptor for hyaluronan-mediated motility (RHAMM). RHAMM is involved in cytoskeletal rearrangement, and recent evidence has suggested that cytoskeleton arrangement and cell shape has a role in macrophage polarization.87 This may be partially mediated by the different molecular weights of HA and their interactions with RHAMM. In endothelial cells, for instance, low molecular weight hyaluronan, but not high molecular weight HAs, were shown to induce endothelial cell proliferation and migration in a RHAMM-dependent manner.88 Complete understanding of hyaluronan binding to RHAMM and subsequent signaling in other cell types has been complicated by the fact that RHAMM does not have a typical trans-membrane domain and is found both intracellularly as well as on the cell surface.89-91 Several HA receptors exist as splice-variants,92 directly interact with one-another,93-94 and bind HA by different mechanisms,92 further obscuring true mechanistic understanding of HA signaling.
Physico-chemical characteristics of HA, dependent on the molecular weight, also convolutes true mechanistic elucidation. Different molecular weights of HA will have varying viscosities and structural conformations in solution.95-97 For instance, short HA chains exhibit non-Gaussian behavior. Flexibility and viscosity of hyaluronan chains increases starting at molecular weights on the order of 104 daltons and reaches a maxima, corresponding to a Gaussian coil on the order of 105 daltons.97 At equimolar concentrations of the HAs, there would be about a 1012 difference in specific viscosity between the highest and lowest molecular weight HA solutions,98-99 which would suggest that viscosity of the HA solutions could dictate the differential phenotypes. However, if viscosity of high (3,000 kDa) and low molecular weight (5 kDa) is adjusted to a similar order of magnitude by modifying the concentration, a similar divergent biologic trend between low and high molecular weight HAs was observed (Figure S3) suggesting that viscosity is not a primary force for polarization following treatment with the HAs. Although also a consequence of dose-response, viscosity’s role in modulating the magnitude of polarization cannot be ignored, as more viscous solutions of either molecular weight elicit exaggerated response (Figure S3). Therefore, although we have categorized the effects by molecular weight, our studies do not exclude the possibility that any one of these characteristics or a combination of them may contribute to the differences we have observed.
Our analysis of hyaluronic acid molecular weights in response to several different activation states offers us preliminary understanding of the role of this polymer in solution. However, our study has analyzed a simplified in vitro model of an immortalized macrophage line analyzed with a limited number of markers. In vivo, there are many different types of macrophages derived from monocytes in the circulating blood or from different tissues, which could interact with the HAs very differently. Additionally, settings in an in vivo environment will include the interaction of other cell types, components of the extracellular matrix, and proteins with these macrophages, which could modify the response that we see under controlled conditions. Although our cell line has been analyzed and confirmed in numerous other studies, there is still the potential that the responses of the immortalized cell line will not correlate to those from a primary cell line, and so further work elucidating the response in these cells as well as a more complex in vivo setting are necessary to truly elucidate the power of varying the molecular weight of HA on macrophage response.
Despite these limitations, our study is the first of its kind to show that molecular weight can modify macrophage phenotype, and that this modification is independent of initial activation state of the macrophage. While it has been recognized that HA plays a role in inflammation, this was thought to be through its scaffolding capabilities allowing cell-matrix interactions.100 Our study provides evidence that HA in solution can modulate the macrophage response, dependent on its molecular weight, inducing or suppressing the expression and production of phenotype-specific genes and products. This understanding may help in the use of hyaluronic acid to control macrophage phenotype, the development of other agents to modify macrophage response, supplement an understanding of the role of extracellular matrix and macrophage response, and improve biomaterial-based systems.
Conclusions
An understanding of macrophage phenotype reprogramming in the context of extracellular matrix biopolymers, such as hyaluronic acid, may begin to clarify improvements in biomaterial based systems and methods of modifying macrophage response to these systems. Our results indicate that hyaluronic acid has molecular weight dependent effects on modulating macrophage phenotype. In solution, the high molecular weight form of the polymer promotes alternative macrophage activation, even when macrophages are challenged with a classically activating stimulus. Conversely, macrophages exposed to low molecular weight HAs are encouraged to produce pro-inflammatory mediators associated with the classically activated state. The most pronounced effects are seen with the lowest and highest molecular weight forms of the polymer. Further mechanistic understanding of HA’s ability to induce macrophage reprogramming is warranted, but several existing studies exhibit results that suggest a high-likelihood of the role of CD44, TLR-4, and RHAMM. We conclude that hyaluronic acid has molecular weight dependent effects on macrophage phenotype that traverse multiple activation states of the macrophage.
Supplementary Material
Acknowledgements
This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant (C06 RR015482) from the National Centre for Research Resources of the National Institutes of Health (NIH). This research has been funded, in part, by the University of Illinois at Chicago Center for Clinical and Translational Science (CCTS) award supported by the NCRR (UL1 TR000050, RAG & JSB), the NIH National Institute for General Medical Studies (R01 GM092850, TJK), the Chicago Biomedical Consortium with support from the Searle funds at the Chicago Community Trust (JER), and a UIC Chancellor’s Graduate Research Fellowship (JER). Additionally, the authors thank Dr. Debra A. Tonetti for use of equipment.
Footnotes
Provisionally accepted to: ACS Biomaterials Science and Engineering (AB-2015-00181r)
Additional experimental procedures and supporting data, specifically influence of endotoxin on macrophage activation, influence of HA concentration on macrophage activation, the levels of markers after 48 and 72 hours of activation, and data presented as the ratios of functional enzymes, nos2 and arg1, interleukins, il12b and il10, and cell surface markers, cd80 and mrc1. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Meyer K, Palmer JW. The Polysaccharide of the Vitreous Humor. J. Biol. Chem. 1934;107:629–634. [Google Scholar]
- 2.Meyer K. The Biological Significance of Hyaluronic Acid and Hyaluronidase. Physiol. Rev. 1947;27:335–359. doi: 10.1152/physrev.1947.27.3.335. [DOI] [PubMed] [Google Scholar]
- 3.Necas J, Bartosikova L, Brauner P, Kolar J. Hyaluronic Acid (Hyaluronan): A Review. Vet. Med. 2008;53:397–411. [Google Scholar]
- 4.Collins MN, Birkinshaw C. Hyaluronic Acid Based Scaffolds for Tissue Engineering-a Review. Carbohyd. Polym. 2013;92:1262–1279. doi: 10.1016/j.carbpol.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 5.Colen S, van den Bekerom MPJ, Mulier M, Haverkamp D. Hyaluronic Acid in the Treatment of Knee Osteoarthritis a Systematic Review and Meta-Analysis with Emphasis on the Efficacy of Different Products. Biodrugs. 2012;26:257–268. doi: 10.2165/11632580-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 6.Beasley KL, Weiss MA, Weiss RA. Hyaluronic Acid Fillers: A Comprehensive Review. Facial Plast. Surg. 2009;25:86–94. doi: 10.1055/s-0029-1220647. [DOI] [PubMed] [Google Scholar]
- 7.Hascall V, Esko JD, Hyaluronan . In: Essentials of Glycobiology. 2 ed. Cummings VA, editor. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2009. Chapter 15. [PubMed] [Google Scholar]
- 8.Stern R, Kogan G, Jedrzejas MJ, Soltes L. The Many Ways to Cleave Hyaluronan. Biotechnol. Adv. 2007;25:537–557. doi: 10.1016/j.biotechadv.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Burdick JA, Prestwich GD. Hyaluronic Acid Hydrogels for Biomedical Applications. Adv. Mater. 2011;23:H41–56. doi: 10.1002/adma.201003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kogan G, Soltes L, Stern R, Gemeiner P. Hyaluronic Acid: A Natural Biopolymer with a Broad Range of Biomedical and Industrial Applications. Biotechnol. Lett. 2007;29:17–25. doi: 10.1007/s10529-006-9219-z. [DOI] [PubMed] [Google Scholar]
- 11.Liu XM, Harmon PS, Maziarz EP, Rah MJ, Merchea MM. Comparative Studies of Hyaluronan in Marketed Ophthalmic Products. Optom. Vis. Sci. 2014;91:32–38. doi: 10.1097/OPX.0000000000000100. [DOI] [PubMed] [Google Scholar]
- 12.Balazs EA. Hyaluronan as an Ophthalmic Viscoelastic Device. Curr. Pharm. Biotechnol. 2008;9:236–238. doi: 10.2174/138920108785161596. [DOI] [PubMed] [Google Scholar]
- 13.Wobig M, Bach G, Beks P, Dickhut A, Runzheimer J, Schwieger G, Vetter G, Balazs E. The Role of Elastoviscosity in the Efficacy of Viscosupplementation for Osteoarthritis of the Knee: A Comparison of Hylan G-F 20 and a Lower-Molecular-Weight Hyaluronan. Clin. Ther. 1999;21:1549–1562. doi: 10.1016/s0149-2918(00)80010-7. [DOI] [PubMed] [Google Scholar]
- 14.Pelletier JP, Martel-Pelletier J. The Pathophysiology of Osteoarthritis and the Implication of the Use of Hyaluronan and Hylan as Therapeutic Agents in Viscosupplementation. J. Rheumatol. 1993;39:S19–S24. [PubMed] [Google Scholar]
- 15.Guidolin D, Franceschi F. Viscosupplementation with High Molecular Weight Native Hyaluronan. Focus on a 1500-2000 Kda Fraction (Hyalubrix(R)) Eur. Rev. Med. Pharmacol. Sci. 2014;18:3326–3338. [PubMed] [Google Scholar]
- 16.Nicoletti G, Ghilardi CG, Scevola S, Faga A. Hyaluronan-Induced Cosmetic Reconstruction of the Nostril. Facial Plast. Surg. 2014;30:81–83. doi: 10.1055/s-0033-1363757. [DOI] [PubMed] [Google Scholar]
- 17.McCracken MS, Khan JA, Wulc AE, Holds JB, Fante RG, Migliori ME, Ebroon DA, Amato MM, Silkiss RZ, Patel BC. Hyaluronic Acid Gel (Restylane) Filler for Facial Rhytids: Lessons Learned from American Society of Ophthalmic Plastic and Reconstructive Surgery Member Treatment of 286 Patients. Ophthal. Plast. Reconstr. Surg. 2006;22:188–191. doi: 10.1097/01.iop.0000217562.64529.ff. [DOI] [PubMed] [Google Scholar]
- 18.Remes A, Williams DF. Immune Response in Biocompatibility. Biomaterials. 1992;13:731–743. doi: 10.1016/0142-9612(92)90010-l. [DOI] [PubMed] [Google Scholar]
- 19.Ialenti A, Di Rosa M. Hyaluronic Acid Modulates Acute and Chronic Inflammation. Agents Actions. 1994;43:44–47. doi: 10.1007/BF02005763. [DOI] [PubMed] [Google Scholar]
- 20.He H, Zhang S, Tighe S, Son J, Tseng SC. Immobilized Heavy Chain-Hyaluronic Acid Polarizes Lipopolysaccharide-Activated Macrophages toward M2 Phenotype. J. Biol. Chem. 2013;288:25792–25803. doi: 10.1074/jbc.M113.479584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuang DM, Wu Y, Chen N, Cheng J, Zhuang SM, Zheng L. Tumor-Derived Hyaluronan Induces Formation of Immunosuppressive Macrophages through Transient Early Activation of Monocytes. Blood. 2007;110:587–595. doi: 10.1182/blood-2007-01-068031. [DOI] [PubMed] [Google Scholar]
- 22.Maharjan AS, Pilling D, Gomer RH. High and Low Molecular Weight Hyaluronic Acid Differentially Regulate Human Fibrocyte Differentiation. PLoS One. 2011;6:e26078. doi: 10.1371/journal.pone.0026078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mytar B, Woloszyn M, Szatanek R, Baj-Krzyworzeka M, Siedlar M, Ruggiero I, Wieckiewicz J, Zembala M. Tumor Cell-Induced Deactivation of Human Monocytes. J. Leukoc. Biol. 2003;74:1094–1101. doi: 10.1189/jlb.0403140. [DOI] [PubMed] [Google Scholar]
- 24.Sokolowska M, Chen LY, Eberlein M, Martinez-Anton A, Liu Y, Alsaaty S, Qi HY, Logun C, Horton M, Shelhamer JH. Low Molecular Weight Hyaluronan Activates Cytosolic Phospholipase A2alpha and Eicosanoid Production in Monocytes and Macrophages. J. Biol. Chem. 2014;289:4470–4488. doi: 10.1074/jbc.M113.515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toole BP. Hyaluronan: From Extracellular Glue to Pericellular Cue. Nat. Rev. Cancer. 2004;4:528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 26.Black KE, Collins SL, Hagan RS, Hamblin MJ, Chan-Li Y, Hallowell RW, Powell JD, Horton MR. Hyaluronan Fragments Induce Ifnbeta Via a Novel Tlr4-Trif-Tbk1-Irf3-Dependent Pathway. J. Inflamm. 2013;10:23. doi: 10.1186/1476-9255-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKee CM, Penno MB, Cowman M, Burdick MD, Strieter RM, Bao C, Noble PW. Hyaluronan (Ha) Fragments Induce Chemokine Gene Expression in Alveolar Macrophages. The Role of Ha Size and Cd44. J. Clin. Invest. 1996;98:2403–2413. doi: 10.1172/JCI119054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Termeer C, Benedix F, Sleeman J, Fieber C, Voith U, Ahrens T, Miyake K, Freudenberg M, Galanos C, Simon JC. Oligosaccharides of Hyaluronan Activate Dendritic Cells Via Toll-Like Receptor 4. J. Exp. Med. 2002;195:99–111. doi: 10.1084/jem.20001858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh P. The Role of Hyaluronic Acid (Hyaluronan) in Health and Disease: Interactions with Cells, Cartilage and Components of Synovial Fluid. Clin. Exp. Rheumatol. 1994;12:75–82. [PubMed] [Google Scholar]
- 30.Garg HG, Hales CA. Chemistry and Biology of Hyaluronan. 1st ed. Elsevier; Boston: 2004. [Google Scholar]
- 31.Fraser JR, Laurent TC, Laurent UB. Hyaluronan: Its Nature, Distribution, Functions and Turnover. J. Intern. Med. 1997;242:27–33. doi: 10.1046/j.1365-2796.1997.00170.x. [DOI] [PubMed] [Google Scholar]
- 32.Mosser DM. The Many Faces of Macrophage Activation. J. Leukocyte Biol. 2003;73:209–212. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 33.Martinez FO, Gordon S. The M1 and M2 Paradigm of Macrophage Activation: Time for Reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosser DM, Edwards JP. Exploring the Full Spectrum of Macrophage Activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosser DM, Zhang X. Activation of Murine Macrophages. Curr. Prot. Immunol. 2008;83:14.2.1–14.2.8. doi: 10.1002/0471142735.im1402s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage Activation and Polarization. Front. Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 37.Stein M, Keshav S, Harris N, Gordon S. Interleukin-4 Potently Enhances Murine Macrophage Mannose Receptor Activity - a Marker of Alternative Immunological Macrophage Activation. J. Exp. Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Odegaard JI, Chawla A. Alternative Macrophage Activation and Metabolism. Annu. Rev. Pathol. 2011;6:275–297. doi: 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon S, Martinez FO. Alternative Activation of Macrophages: Mechanism and Functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Gordon S. Alternative Activation of Macrophages. Nat. Rev. Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 41.Luo Y, Prestwich GD. Synthesis and Selective Cytotoxicity of a Hyaluronic Acid-Antitumor Bioconjugate. Bioconjug. Chem. 1999;10:755–763. doi: 10.1021/bc9900338. [DOI] [PubMed] [Google Scholar]
- 42.Schiller J, Arnhold J, Benard S, Reichl S, Arnold K. Cartilage Degradation by Hyaluronate Lyase and Chondroitin Abc Lyase: A Maldi-Tof Mass Spectrometric Study. Carbohydr. Res. 1999;318:116–122. doi: 10.1016/s0008-6215(99)00063-4. [DOI] [PubMed] [Google Scholar]
- 43.Yeung B, Marecak D. Molecular Weight Determination of Hyaluronic Acid by Gel Filtration Chromatography Coupled to Matrix-Assisted Laser Desorption Ionization Mass Spectrometry. J. Chromatogr. A. 1999;852:573–581. doi: 10.1016/s0021-9673(99)00647-0. [DOI] [PubMed] [Google Scholar]
- 44.Tezel A, Fredrickson GH. The Science of Hyaluronic Acid Dermal Fillers. J. Cosmet. Laser Ther. 2008;10:35–42. doi: 10.1080/14764170701774901. [DOI] [PubMed] [Google Scholar]
- 45.Bellamy N, Campbell J, Robinson V, Gee T, Bourne R, Wells G. Viscosupplementation for the Treatment of Osteoarthritis of the Knee. Cochrane Database Syst. Rev. 2006 doi: 10.1002/14651858.CD005321.pub2. CD005321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo GH, LaValley M, McAlindon T, Felson DT. Intra-Articular Hyaluronic Acid in Treatment of Knee Osteoarthritis: A Meta-Analysis. JAMA, J. Am. Med. Assoc. 2003;290:3115–3121. doi: 10.1001/jama.290.23.3115. [DOI] [PubMed] [Google Scholar]
- 47.Laurent TC, Fraser JR. The Properties and Turnover of Hyaluronan. Ciba Found. Symp. 1986;124:9–29. doi: 10.1002/9780470513385.ch2. [DOI] [PubMed] [Google Scholar]
- 48.Laurent UB. Hyaluronate in Aqueous Humour. Exp. Eye Res. 1981;33:147–155. doi: 10.1016/s0014-4835(81)80063-2. [DOI] [PubMed] [Google Scholar]
- 49.Pettersson T, Froseth B, Riska H, Klockars M. Concentration of Hyaluronic Acid in Pleural Fluid as a Diagnostic Aid for Malignant Mesothelioma. Chest. 1988;94:1037–1039. doi: 10.1378/chest.94.5.1037. [DOI] [PubMed] [Google Scholar]
- 50.Balazs EA, Watson D, Duff IF, Roseman S. Hyaluronic Acid in Synovial Fluid. I. Molecular Parameters of Hyaluronic Acid in Normal and Arthritis Human Fluids. Arthritis Rheum. 1967;10:357–376. doi: 10.1002/art.1780100407. [DOI] [PubMed] [Google Scholar]
- 51.Ralph P, Moore MA, Nilsson K. Lysozyme Synthesis by Established Human and Murine Histiocytic Lymphoma Cell Lines. J. Exp. Med. 1976;143:1528–1533. doi: 10.1084/jem.143.6.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ralph P, Nakoinz I. Antibody-Dependent Killing of Erythrocyte and Tumor Targets by Macrophage-Related Cell Lines: Enhancement by Ppd and Lps. J. Immunol. 1977;119:950–954. [PubMed] [Google Scholar]
- 53.Iyamu EW, Asakura T, Woods GM. A Colorimetric Microplate Assay Method for High-Throughput Analysis of Arginase Activity in Vitro. Anal. Biochem. 2008;383:332–334. doi: 10.1016/j.ab.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morris SM, Kepka-Lenhart D, Chen LC. Differential Regulation of Arginases and Inducible Nitric Oxide Synthase in Murine Macrophage Cells. Am. J. Physiol.: Endocrinol. Metab. 1998;275:E740–E747. doi: 10.1152/ajpendo.1998.275.5.E740. [DOI] [PubMed] [Google Scholar]
- 55.Schenborn E, Groskreutz D. Reporter Gene Vectors and Assays. Mol. Biotechnol. 1999;13:29–44. doi: 10.1385/MB:13:1:29. [DOI] [PubMed] [Google Scholar]
- 56.Schmittgen TD, Livak KJ. Analyzing Real-Time Pcr Data by the Comparative C(T) Method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 57.Gordon S. Macrophage Heterogeneity and Tissue Lipids. J. Clin. Invest. 2007;117:89–93. doi: 10.1172/JCI30992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-Like Receptor-4 Mediates Lipopolysaccharide-Induced Signal Transduction. J. Biol. Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 59.Iwamoto M, Kurachi M, Nakashima T, Kim D, Yamaguchi K, Oda T, Iwamoto Y, Muramatsu T. Structure-Activity Relationship of Alginate Oligosaccharides in the Induction of Cytokine Production from Raw264.7 Cells. Febs Lett. 2005;579:4423–4429. doi: 10.1016/j.febslet.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 60.Rogers TH, Babensee JE. Altered Adherent Leukocyte Profile on Biornaterials in Toll-Like Receptor 4 Deficient Mice. Biomaterials. 2010;31:594–601. doi: 10.1016/j.biomaterials.2009.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grandjean-Laquerriere A, Tabary O, Jacquot J, Richard D, Frayssinet P, Guenounou M, Laurent-Maquin D, Laquerriere P, Gangloff S. Involvement of Toll-Like Receptor 4 in the Inflammatory Reaction Induced by Hydroxyapatite Particles. Biomaterials. 2007;28:400–404. doi: 10.1016/j.biomaterials.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 62.Campo GM, Avenoso A, Campo S, D’Ascola A, Traina P, Rugolo CA, Calatroni A. Differential Effect of Molecular Mass Hyaluronan on Lipopolysaccharide-Induced Damage in Chondrocytes. Innate Immun. 2010;16:48–63. doi: 10.1177/1753425909340419. [DOI] [PubMed] [Google Scholar]
- 63.Hu X, Ivashkiv LB. Cross-Regulation of Signaling Pathways by Interferon-Gamma: Implications for Immune Responses and Autoimmune Diseases. Immunity. 2009;31:539–50. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The Chemokine System in Diverse Forms of Macrophage Activation and Polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 65.Porcheray F, Viaud S, Rimaniol A-C, Leone C, Samah B, Dereuddre-Bosquet N, Dormont D, Gras G. Macrophage Activation Switching: An Asset for the Resolution of Inflammation. Clin. Exp. Immunol. 2005;142:481–489. doi: 10.1111/j.1365-2249.2005.02934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Noble PW. Hyaluronan and Its Catabolic Products in Tissue Injury and Repair. Matrix Biol. 2002;21:25–29. doi: 10.1016/s0945-053x(01)00184-6. [DOI] [PubMed] [Google Scholar]
- 67.Chen WYJ, Abatangelo G. Functions of Hyaluronan in Wound Repair. Wound Repair Regen. 1999;7:79–89. doi: 10.1046/j.1524-475x.1999.00079.x. [DOI] [PubMed] [Google Scholar]
- 68.Oksala O, Salo T, Tammi R, Hakkinen L, Jalkanen M, Inki P, Larjava H. Expression of Proteoglycans and Hyaluronan During Wound-Healing. J. Histochem. Cytochem. 1995;43:125–135. doi: 10.1177/43.2.7529785. [DOI] [PubMed] [Google Scholar]
- 69.Ponta H, Sherman L, Herrlich PA. Cd44: From Adhesion Molecules to Signalling Regulators. Nat. Rev. Mol. Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 70.Gee K, Lim W, Ma W, Nandan D, Diaz-Mitoma F, Kozlowski M, Kumar A. Differential Regulation of Cd44 Expression by Lipopolysaccharide (Lps) and Tnf-Alpha in Human Monocytic Cells: Distinct Involvement of C-Jun N-Terminal Kinase in Lps-Induced Cd44 Expression. J. Immunol. 2002;169:5660–5672. doi: 10.4049/jimmunol.169.10.5660. [DOI] [PubMed] [Google Scholar]
- 71.Hodge-Dufour J, Noble PW, Horton MR, Bao C, Wysoka M, Burdick MD, Strieter RM, Trinchieri G, Pure E. Induction of Il-12 and Chemokines by Hyaluronan Requires Adhesion-Dependent Priming of Resident but Not Elicited Macrophages. J. Immunol. 1997;159:2492–2500. [PubMed] [Google Scholar]
- 72.Levesque MC, Haynes BF. Cytokine Induction of the Ability of Human Monocyte Cd44 to Bind Hyaluronan Is Mediated Primarily by Tnf-Alpha and Is Inhibited by Il-4 and Il-13. J. Immunol. 1997;159:6184–6194. [PubMed] [Google Scholar]
- 73.Underhill CB, Nguyen HA, Shizari M, Culty M. Cd44 Positive Macrophages Take up Hyaluronan During Lung Development. Dev. Biol. 1993;155:324–336. doi: 10.1006/dbio.1993.1032. [DOI] [PubMed] [Google Scholar]
- 74.Levesque MC, Haynes BF. Tnf Alpha and Il-4 Regulation of Hyaluronan Binding to Monocyte Cd44 Involves Posttranslational Modification of Cd44. Cell Immunol. 1999;193:209–218. doi: 10.1006/cimm.1999.1456. [DOI] [PubMed] [Google Scholar]
- 75.Brown KL, Maiti A, Johnson P. Role of Sulfation in Cd44-Mediated Hyaluronan Binding Induced by Inflammatory Mediators in Human Cd14(+) Peripheral Blood Monocytes. J. Immunol. 2001;167:5367–5374. doi: 10.4049/jimmunol.167.9.5367. [DOI] [PubMed] [Google Scholar]
- 76.Pure E, Cuff CA. A Crucial Role for Cd44 in Inflammation. Trends Mol. Med. 2001;7:213–221. doi: 10.1016/s1471-4914(01)01963-3. [DOI] [PubMed] [Google Scholar]
- 77.Bollyky PL, Lord JD, Masewicz SA, Evanko SP, Buckner JH, Wight TN, Nepom GT. Cutting Edge: High Molecular Weight Hyaluronan Promotes the Suppressive Effects of Cd4+Cd25+ Regulatory T Cells. J. Immunol. 2007;179:744–747. doi: 10.4049/jimmunol.179.2.744. [DOI] [PubMed] [Google Scholar]
- 78.Firan M, Dhillon S, Estess P, Siegelman MH. Suppressor Activity and Potency among Regulatory T Cells Is Discriminated by Functionally Active Cd44. Blood. 2006;107:619–627. doi: 10.1182/blood-2005-06-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manrique SZ, Correa MA, Hoelzinger DB, Dominguez AL, Mirza N, Lin HH, Stein-Streilein J, Gordon S, Lustgarten J. Foxp3-Positive Macrophages Display Immunosuppressive Properties and Promote Tumor Growth. J. Exp. Med. 2011;208:1485–1499. doi: 10.1084/jem.20100730. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Hart SP, Rossi AG, Haslett C, Dransfield I. Characterization of the Effects of Cross-Linking of Macrophage Cd44 Associated with Increased Phagocytosis of Apoptotic Pmn. PLoS One. 2012;7:e33142. doi: 10.1371/journal.pone.0033142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vivers S, Dransfield I, Hart SP. Role of Macrophage Cd44 in the Disposal of Inflammatory Cell Corpses. Clin. Sci. 2002;103:441–449. doi: 10.1042/cs1030441. [DOI] [PubMed] [Google Scholar]
- 82.Kaisho T, Akira S. Toll-Like Receptors and Their Signaling Mechanism in Innate Immunity. Acta Odontol. Scand. 2001;59:124–130. doi: 10.1080/000163501750266701. [DOI] [PubMed] [Google Scholar]
- 83.Medzhitov R. Toll-Like Receptors and Innate Immunity. Nat. Rev. Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 84.Akira S, Takeda K. Toll-Like Receptor Signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 85.Hume DA, Underhill DM, Sweet MJ, Ozinsky AO, Liew FY, Aderem A. Macrophages Exposed Continuously to Lipopolysaccharide and Other Agonists That Act Via Toll-Like Receptors Exhibit a Sustained and Additive Activation State. BMC Immunol. 2001;2:11. doi: 10.1186/1471-2172-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Campo GM, Avenoso A, Campo S, D’Ascola A, Nastasi G, Calatroni A. Molecular Size Hyaluronan Differently Modulates Toll-Like Receptor-4 in Lps-Induced Inflammation in Mouse Chondrocytes. Biochimie. 2010;92:204–215. doi: 10.1016/j.biochi.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 87.McWhorter FY, Wang T, Nguyen P, Chung T, Liu WF. Modulation of Macrophage Phenotype by Cell Shape. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17253–17258. doi: 10.1073/pnas.1308887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matou-Nasri S, Gaffney J, Kumar S, Slevin M. Oligosaccharides of Hyaluronan Induce Angiogenesis through Distinct Cd44 and Rhamm-Mediated Signalling Pathways Involving Cdc2 and Gamma-Adducin. Int. J. Oncol. 2009;35:761–773. doi: 10.3892/ijo_00000389. [DOI] [PubMed] [Google Scholar]
- 89.Hardwick C, Hoare K, Owens R, Hohn HP, Hook M, Moore D, Cripps V, Austen L, Nance DM, Turley EA. Molecular-Cloning of a Novel Hyaluronan Receptor That Mediates Tumor-Cell Motility. J. Cell Biol. 1992;117:1343–1350. doi: 10.1083/jcb.117.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hofmann M, Fieber C, Assmann V, Gottlicher M, Sleeman J, Plug R, Howells N, von Stein O, Ponta H, Herrlich P. Identification of Ihabp, a 95 Kda Intracellular Hyaluronate Binding Protein. J. Cell Sci. 1998;111(Pt 12):1673–1684. doi: 10.1242/jcs.111.12.1673. [DOI] [PubMed] [Google Scholar]
- 91.Zhang S, Chang MC, Zylka D, Turley S, Harrison R, Turley EA. The Hyaluronan Receptor Rhamm Regulates Extracellular-Regulated Kinase. J. Biol. Chem. 1998;273:11342–11348. doi: 10.1074/jbc.273.18.11342. [DOI] [PubMed] [Google Scholar]
- 92.Entwistle J, Hall CL, Turley EA. Receptors: Regulators of Signalling to the Cytoskeleton. J. Cell Biochem. 1996;61:569–577. doi: 10.1002/(sici)1097-4644(19960616)61:4<569::aid-jcb10>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 93.Nikitovic D, Kouvidi K, Karamanos NK, Tzanakakis GN. The Roles of Hyaluronan/Rhamm/Cd44 and Their Respective Interactions Along the Insidious Pathways of Fibrosarcoma Progression. Biomed. Res. Int. 2013;2013:929531. doi: 10.1155/2013/929531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taylor KR, Yamasaki K, Radek KA, Di Nardo A, Goodarzi H, Golenbock D, Beutler B, Gallo RL. Recognition of Hyaluronan Released in Sterile Injury Involves a Unique Receptor Complex Dependent on Toll-Like Receptor 4, Cd44, and Md-2. J. Biol. Chem. 2007;282:18265–18275. doi: 10.1074/jbc.M606352200. [DOI] [PubMed] [Google Scholar]
- 95.Takahashi R, Kubota K, Kawada M, Okamoto A. Effect of Molecular Weight Distribution on the Solution Properties of Sodium Hyaluronate in 0.2m Nacl Solution. Biopolymers. 1999;50:87–98. [Google Scholar]
- 96.Gribbon P, Heng BC, Hardingham TE. The Analysis of Intermolecular Interactions in Concentrated Hyaluronan Solutions Suggest No Evidence for Chain-Chain Association. Biochem. J. 2000;350(Pt 1):329–335. [PMC free article] [PubMed] [Google Scholar]
- 97.Shimada E, Matsumura G. Viscosity and Molecular-Weight of Hyaluronic Acids. J. Biochem. 1975;78:513–517. doi: 10.1093/oxfordjournals.jbchem.a130935. [DOI] [PubMed] [Google Scholar]
- 98.Matsuoka S, Cowman MK. Equation of State for Polymer Solution. Polymer. 2002;43:3447–3453. [Google Scholar]
- 99.Mendichi R, Soltes L, Giacometti Schieroni A. Evaluation of Radius of Gyration and Intrinsic Viscosity Molar Mass Dependence and Stiffness of Hyaluronan. Biomacromolecules. 2003;4:1805–1810. doi: 10.1021/bm0342178. [DOI] [PubMed] [Google Scholar]
- 100.Laurent TC, Fraser JR, Hyaluronan FASEB J. 1992;6:2397–2404. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.