Figure 1.
Kinetochore Proteins Show Single-Peak Localization on Each Chromosome of N. castellii
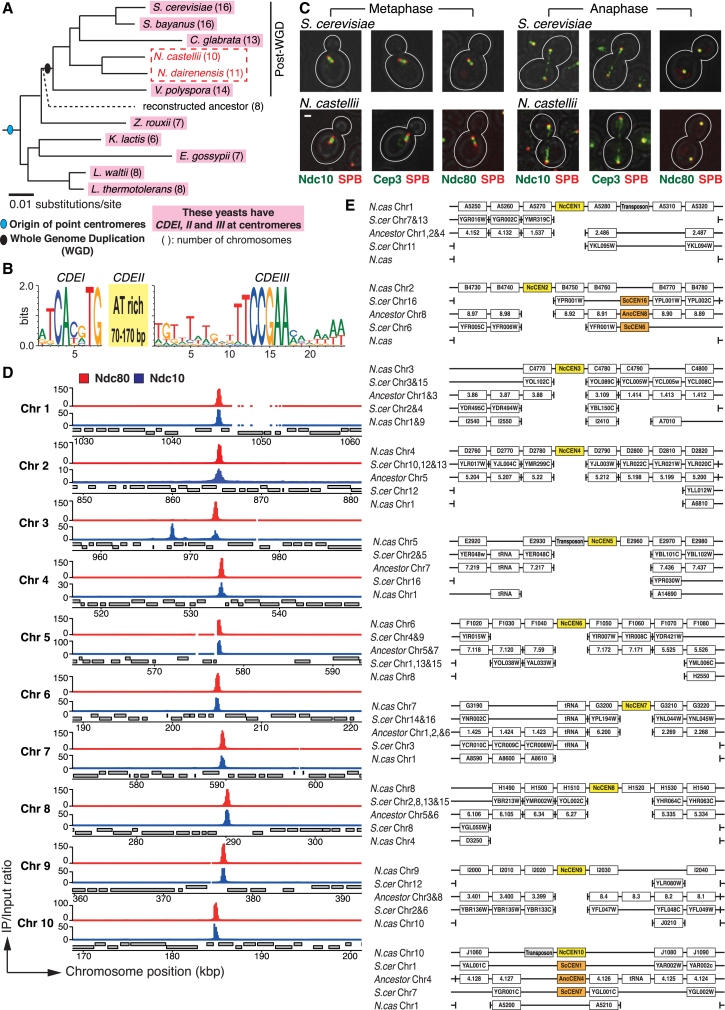
(A) A clade of budding yeast species carries point centromeres with common consensus DNA sequences CDEI, II, and III (pink), except for N. castellii and N. dairenensis (red rectangle). Phylogenetic tree of budding yeast species is taken from [6, 7] with modification. Horizontal length in the tree is proportional to the change in genomic DNA sequences. The WGD (black oval) is estimated to have occurred approximately 100 million years ago [8], and the N. castellii/N. dairenensis divergence is about half this age.
(B) Nucleotide sequence of CDEI, II, and III [5, 6]. Logos of nucleotides graphically represent their frequency at individual positions. CDEII is 70–170 bp DNA sequence with >79% AT content.
(C) Localization of Ndc10, Cep3, and Ndc80 in representative S. cerevisiae and N. castellii cells (in metaphase and anaphase). Ndc10, Cep3, and Ndc80 were tagged with three tandem copies of GFP (3× GFP) in N. castellii (T11587, T11584, and T11586, respectively) and with 1× GFP in S. cerevisiae (T11520, T11522, and T11521, respectively). Spindle pole body (SPB) components, Spc42 and Spc110, were tagged with 4× mCherry and 1× mCherry in N. castellii and S. cerevisiae, respectively. Cell shapes are outlined in white. The scale bar represents 1 μm. Metaphase and anaphase are defined by the distance between two SPBs (<2.5 μm and >3 μm, respectively). Note that, during anaphase, CBF3 also localizes on the spindle as well as at kinetochores [9].
(D) Ndc80 (red) shows single-peak localization on each chromosome (Chr) of N. castellii. Ndc10 (blue) also shows peaks at the same regions. NDC80-6xHA (T9328) and NDC10-6xHA (T9326) cells were processed for ChIP-seq. Gray bars represent open reading frames of genes; top on Watson strand, bottom on Crick strand. Chromosome positions are shown in length (kilo base pairs [kbp]) from the left telomere. In addition to the positions of Ndc80 peaks, Ndc10 showed two extra peaks (Figure S1A); one of them is at 968 kbp on chromosome 3, as shown here.
(E) Gene order in S. cerevisiae and the reconstructed ancestor, aligned around N. castellii CENs (yellow box). Gene orders were analyzed using YGOB [10]. Vertical tick bars represent gaps; i.e., genes to the right and left are not neighbors on a chromosome. Two chromosome series of N. castellii and S. cerevisiae are aligned with one series of the ancestor because N. castellii and S. cerevisiae are post-WGD yeasts (see A). Orange boxes represent centromeres in the ancestor (AncCEN) and S. cerevisiae (ScCEN).
See also Figure S1.