Figure 2.
N. castellii Candidate Centromeres Show Dynamic Localizations during the Cell Cycle, which Are Consistent with Those of Functional Centromeres
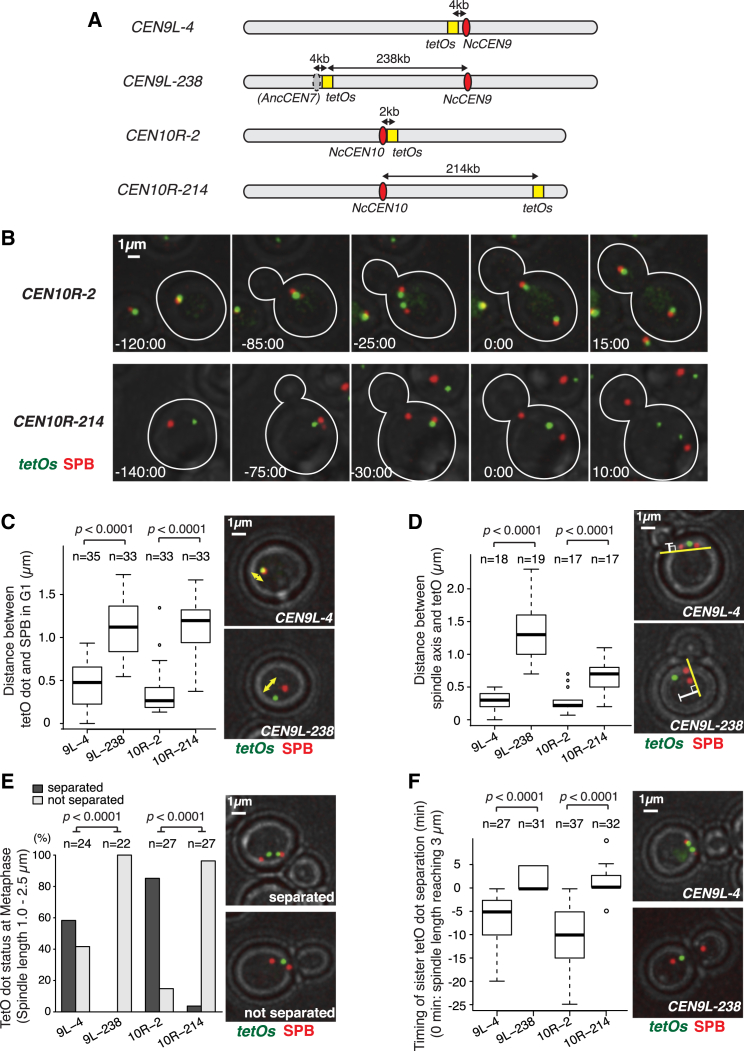
(A) Diagram showing positions of the insertion of tetO arrays (112 tandem repeats; yellow box) on chromosomes 9 and 10. NcCEN9 and NcCEN10 are shown as red ovals. The corresponding position of the ancestor CEN7 (AncCEN7), based on synteny, is shown as a gray oval.
(B) Live-cell images of tetO arrays shown in (A). Images of SPC42-4xmCherry TetR-GFP cells with tetOx112 at CEN10R-2 (T11466) or at CEN10R-214 (T11467) were acquired every 5 min. Spc42 is an SPB component. Time (min: s) is shown relative to anaphase onset (when the distance between SPBs exceeded 3 μm). The scale bar represents 1 μm.
(C–F) Analyses of tetOs localization in cells. Live-cell images of SPC42-4xmCherry TetR-GFP cells with tetOx112 at CEN9L-4 (T11501), CEN9L-238 (T11500), CEN10R-2 (T11466), or CEN10R-214 (T11467) were acquired every 5 min. (C) Distance between the SPB and the tetO dot in G1 phase is shown (in cells with one SPB but no bud). In box plots, a thick line represents a median; a box shows the range of the first to third quartile (interquartile range: IQR); the upper and lower whiskers show the maximum and minimum values, respectively, which do not exceed 3/2 IQR beyond the box; and open circles show outliers. (D) Distance between the tetO dot and the spindle axis in metaphase is shown (cells with two SPBs less than 2 μm apart and with a single tetO dot). Box plots are as in (C). (E) Frequency of separation and non-separation of the tetO dot in metaphase is shown (cells with two SPBs 1.0–2.5 μm apart). (F) Timing of separation of the tetO dot, relative to anaphase onset, is shown (as defined in B). Representative cells show separation of sister tetO dots (at CEN9L-4) before anaphase onset (top) and no separation of them (at CEN9-L-238) after anaphase onset (bottom). Box plots are as in (C).