Figure 4.
Consensus DNA Elements in N. castellii CENs Are Important for the Centromere Function in the Context of Authentic Chromosomes
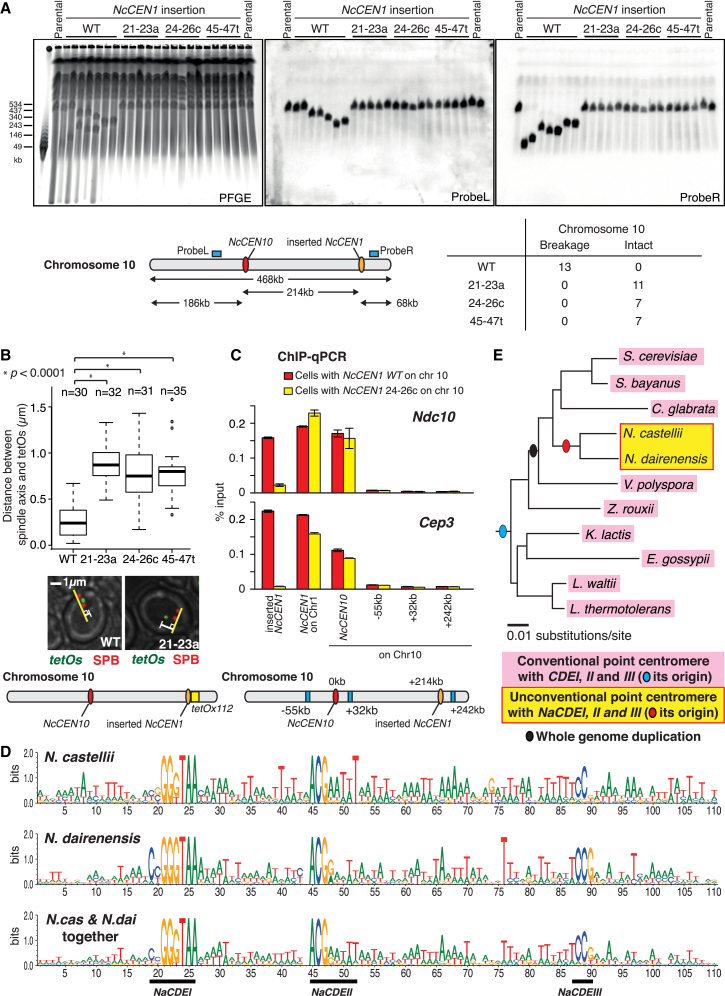
(A) NcCEN1 wild-type (WT; 1,173 bp; Figure 3B) and its mutants (1,173 bp with 21–23a, 24–26c, and 45–47t; Figure 3C) were inserted at 214 kb right of NcCEN10 on chromosome 10 (orange oval in diagram) in haploid N. castellii cells (T11605). Karyotypes of individual clones were analyzed (representative examples are shown here) by pulsed field gel electrophoresis (PFGE), followed by Southern blots with ProbeL and ProbeR; see positions of the probes in diagram. Table shows the number of clones that did, or did not, show breakage of chromosome 10 (shortening detected by ProbeL and/or ProbeR; Figure S3A).
(B) NcCEN1 wild-type (WT; T11632), 21–23a (Figure 3C; T11842), 24–26c (T11843), and 45–47t (T11844), each marked with tetOx112, were inserted at 214 kb right of NcCEN10 on chromosome 10 in SPC42-4xmCherry TetR-GFP cells. Cells were imaged, and the distance between the spindle axis and inserted NcCEN1 was analyzed as in Figure 2D. Box plots are as in Figure 2C.
(C) NDC10-3xFLAG cells with inserted NcCEN1 wild-type (WT; T11845) and 24–26c (T11846), as well as CEP3-3xFLAG cells with inserted NcCEN1 wild-type (WT; T11847) and 24–26c (T11848), were processed for ChIP-qPCR. Diagram shows chromosome loci for quantification by PCR (blue box). Primers for qPCR were designed to distinguish the original NcCEN1 on chromosome 1 and NcCEN1 inserted on chromosome 10. Error bars represent SEM (n = 3).
(D) N. dairenensis candidate centromeres (Figure S4B) have consensus DNA elements, NaCDEI, II, and III, which are also found at N. castellii CENs (Figure 3A). Logos of nucleotides graphically represent their frequency at individual positions, in aligned N. castellii centromeres (top), N. dairenensis candidate centromeres (middle), and both together (bottom).
(E) Conclusion of this study: budding yeasts, highlighted in pink, have point centromeres with CDEI, II, and III, which are conserved across several species. In contrast, N. castellii and N. dairenensis, highlighted in yellow, carry unconventional point centromeres with NaCDEI, II, and III, which have very different DNA sequences from conventional CDEI, II, and III. Blue and red ovals show the origins of the two types of point centromere in the yeast phylogenetic tree.
See also Figures S3 and S4.